Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase - PubMed (original) (raw)
Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase
Michelle L Ebbs et al. Plant Cell. 2006 May.
Abstract
In Arabidopsis thaliana, heterochromatin formation is guided by double-stranded RNA (dsRNA), which triggers methylation of histone H3 at Lys-9 (H3 mK9) and CG plus non-CG methylation on identical DNA sequences. At heterochromatin targets including transposons and centromere repeats, H3 mK9 mediated by the Su(var)3-9 homologue 4 (SUVH4)/KYP histone methyltransferase (MTase) is required for the maintenance of non-CG methylation by the CMT3 DNA MTase. Here, we show that although SUVH4 is the major H3 K9 MTase, the SUVH5 protein also has histone MTase activity in vitro and contributes to the maintenance of H3 mK9 and CMT3-mediated non-CG methylation in vivo. Strikingly, the relative contributions of SUVH4, SUVH5, and a third related histone MTase, SUVH6, to non-CG methylation are locus-specific. For example, SUVH4 and SUVH5 together control transposon sequences with only a minor contribution from SUVH6, whereas SUVH4 and SUVH6 together control a transcribed inverted repeat source of dsRNA with only a minor contribution from SUVH5. This locus-specific variation suggests different mechanisms for recruiting or activating SUVH enzymes at different heterochromatic sequences. The suvh4 suvh5 suvh6 triple mutant loses both monomethyl and dimethyl H3 K9 at target loci. The suvh4 suvh5 suvh6 mutant also displays a loss of non-CG methylation similar to a cmt3 mutant, indicating that SUVH4, SUVH5, and SUVH6 together control CMT3 activity.
Figures
Figure 1.
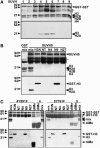
SUVH5 Has in Vitro Histone MTase Activity. Coomassie blue–stained gels (top panels) and their fluorograms (bottom panels) are shown. GST-SUVH indicates the positions of full-length recombinant SUVH proteins, and asterisks indicate GST-SUVH truncations. Positions of protein molecular mass markers in kilodaltons are shown at left. (A) In vitro histone MTase assays were performed with a whole histone mix and each of the nine GST-tagged SUVH proteins. (B) Specificity of SUVH5 for H3 K9. GST-SUVH5 was incubated with a bovine histone mix or a GST-H3 N-terminal peptide as indicated: H3N is the wild type, N4 has K9 and K27 mutated to Arg, N9 has K4 and K27 mutated to Arg, N27 has K4 and K9 mutated to Arg, and NT has K4, K9 and K27 mutated to Arg (Tachibana et al., 2001). GST alone was incubated with the histone mix as a control. (C) Tyr-to-Phe mutations convert SUVH4 or SUVH5 from an H3 K9 mono-/di-MTase to an H3 K9 tri-MTase. GST-SUVH4Y591F, GST-SUVH4, GST-SUVH5Y761F, and GST-SUVH5 were assayed for activity on bovine histone mix (mix), GST-H3 peptides, or a biotinylated H3 K9 dimethyl peptide (diMe).
Figure 2.
The suvh4 suvh5 suvh6 Mutant Shows Similar PAI2 Transcriptional Reactivation to a cmt3 Mutant in the pai1 Reporter Background. Representative 2-week-old seedlings are shown photographed under visible light (right row) or short-wave UV light (left row). All mutations assayed were in the Ws pai1 background, with WT indicating wild type, 4 indicating suvh4R302*, 5 indicating suvh5-1, 6 indicating suvh6-1, and cmt3 indicating cmt3illa (Bartee et al., 2001).
Figure 3.
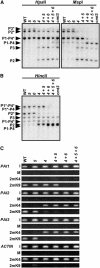
SUVH4, SUVH5, and SUVH6 All Contribute to PAI1-PAI4 Non-CG Methylation. (A) and (B) DNA gel blot assays for PAI DNA methylation patterning. Genomic DNA from the indicated mutants was cleaved with _Hpa_II or _Msp_I isoschizomers (A) or _Hin_cII (B) and used in DNA gel blot analysis with a PAI1 cDNA probe. P1-P4 indicates PAI1-PAI4, P2 indicates PAI2, and P3 indicates PAI3, with bands diagnostic of methylation on _PAI_-internal sites denoted with asterisks. (C) ChIP analysis of H3 2mK4 and H3 2mK9 patterning on the PAI genes. Primer sets specific for PAI1-PAI4 (indicated as PAI1), PAI2, PAI3, or ACTIN were used to amplify PCR products from total input chromatin (I), no-antibody mock precipitation control (M), chromatin immunoprecipitated with H3 anti-dimethyl K4 antibodies (2mK4), or chromatin immunoprecipitated with H3 anti-dimethyl K9 antibodies (2mK9) from the indicated mutants. GelStar-stained PCR products are shown. These results were reproduced in three independent experiments (see Supplemental Figure 3 online), with a representative data set shown. ACTIN is a control unmethylated transcribed gene previously determined to be enriched for H3 2mK4 but not H3 2mK9 in wild-type and DNA methylation–deficient mutant backgrounds (Johnson et al., 2002; Ebbs et al., 2005). Genotypes are as described for Figure 2.
Figure 4.
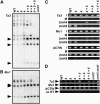
SUVH5 Contributes to Ta3 and Mu1 Transposon Non-CG Methylation and Ta3 Transcriptional Silencing in the Absence of SUVH4. (A) and (B) DNA gel blot assays for transposon DNA methylation patterning. DNA from the indicated mutants was cleaved with _Msp_I (A) or _Hin_dIII plus _Msp_I (B) and used in DNA gel blot analysis with a Ta3 probe (A) or an Mu1 probe (B). Arrowheads at left indicate the positions of fully cleaved bands. (C) ChIP analysis of H3 2mK4 and H3 2mK9 patterning on transposons. Primer sets specific for Ta3, Mu1, or ACTIN were used to amplify PCR products from total input chromatin (I), no-antibody mock precipitation control (M), chromatin immunoprecipitated with H3 anti-dimethyl K4 antibodies (2mK4), or chromatin immunoprecipitated with H3 anti-dimethyl K9 antibodies (2mK9) from the indicated mutants. GelStar-stained PCR products are shown. These results were reproduced in three independent experiments (see Supplemental Figure 3 online), with a representative data set shown. (D) Semiquantitative RT-PCR analysis of transposon transcription. Primer sets specific for Ta3, Mu1, or ACTIN were used for RT-PCR analysis of total RNA prepared from 3-week-old plants of the indicated genotypes. The no-RT control is shown for the ACTIN primer set. Genotypes are as described for Figure 2. The cmt3 met1 mutant (Ebbs et al., 2005) was used as a control for RT-PCR analysis.
Figure 5.
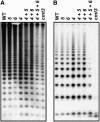
SUVH4, SUVH5, and SUVH6 Together Control the Majority of CMT3-Mediated DNA Methylation at Repetitive Sequences. DNA gel blot assays for repetitive sequences. DNA from the indicated mutants was cleaved with _Msp_I and used in DNA gel blot analysis with a 180-bp centromere repeat probe (A) or a 5S rDNA probe (B). Genotypes are as described for Figure 2.
Figure 6.
SUVH4, SUVH5, and SUVH6 Together Control H3 1mK9. ChIP analysis of H3 1mK9 patterning on Ta3 and Mu1 transposons (A), the PAI1-PAI4 transcribed inverted repeat (indicated as PAI1) (B), the silenced PAI2 and PAI3 genes (C), and the unmethylated transcribed control gene ACTIN (D). Primer sets specific for each gene were used to amplify PCR products from total input chromatin (I), no-antibody mock precipitation control (M), or chromatin immunoprecipitated with H3 anti-monomethyl K9 antibodies (1mK9) from the indicated mutants. GelStar-stained PCR products are shown. These results were reproduced in three independent experiments (see Supplemental Figure 6 online), with a representative data set shown. Genotypes are as described for Figure 2.
Similar articles
- H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases.
Ebbs ML, Bartee L, Bender J. Ebbs ML, et al. Mol Cell Biol. 2005 Dec;25(23):10507-15. doi: 10.1128/MCB.25.23.10507-10515.2005. Mol Cell Biol. 2005. PMID: 16287862 Free PMC article. - HISTONE DEACETYLASE6 Acts in Concert with Histone Methyltransferases SUVH4, SUVH5, and SUVH6 to Regulate Transposon Silencing.
Yu CW, Tai R, Wang SC, Yang P, Luo M, Yang S, Cheng K, Wang WC, Cheng YS, Wu K. Yu CW, et al. Plant Cell. 2017 Aug;29(8):1970-1983. doi: 10.1105/tpc.16.00570. Epub 2017 Aug 4. Plant Cell. 2017. PMID: 28778955 Free PMC article. - Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase.
Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Jackson JP, et al. Nature. 2002 Apr 4;416(6880):556-60. doi: 10.1038/nature731. Epub 2002 Mar 17. Nature. 2002. PMID: 11898023 - Heterochromatin proteins and the control of heterochromatic gene silencing in Arabidopsis.
Fischer A, Hofmann I, Naumann K, Reuter G. Fischer A, et al. J Plant Physiol. 2006 Feb;163(3):358-68. doi: 10.1016/j.jplph.2005.10.015. Epub 2005 Dec 27. J Plant Physiol. 2006. PMID: 16384625 Review. - Chromatin-based silencing mechanisms.
Bender J. Bender J. Curr Opin Plant Biol. 2004 Oct;7(5):521-6. doi: 10.1016/j.pbi.2004.07.003. Curr Opin Plant Biol. 2004. PMID: 15337094 Review.
Cited by
- Mutations in EDM2 selectively affect silencing states of transposons and induce plant developmental plasticity.
Tsuchiya T, Eulgem T. Tsuchiya T, et al. Sci Rep. 2013;3:1701. doi: 10.1038/srep01701. Sci Rep. 2013. PMID: 23609044 Free PMC article. - Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome.
Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Stroud H, et al. Cell. 2013 Jan 17;152(1-2):352-64. doi: 10.1016/j.cell.2012.10.054. Epub 2013 Jan 11. Cell. 2013. PMID: 23313553 Free PMC article. - Epigenetic Modifications during Angiosperm Gametogenesis.
Migicovsky Z, Kovalchuk I. Migicovsky Z, et al. Front Plant Sci. 2012 Feb 6;3:20. doi: 10.3389/fpls.2012.00020. eCollection 2012. Front Plant Sci. 2012. PMID: 22645573 Free PMC article. - Diversity and dynamics of DNA methylation: epigenomic resources and tools for crop breeding.
Kawakatsu T, Ecker JR. Kawakatsu T, et al. Breed Sci. 2019 Jun;69(2):191-204. doi: 10.1270/jsbbs.19005. Epub 2019 May 14. Breed Sci. 2019. PMID: 31481828 Free PMC article. Review. - Differential epigenetic regulation within an Arabidopsis retroposon family.
Rangwala SH, Richards EJ. Rangwala SH, et al. Genetics. 2007 May;176(1):151-60. doi: 10.1534/genetics.107.071092. Epub 2007 Mar 4. Genetics. 2007. PMID: 17339215 Free PMC article.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
- Baumbusch, L.O., Thorstensen, T., Krauss, V., Fischer, A., Naumann, K., Assalkhou, R., Schulz, I., Reuter, G., and Aalen, R.B. (2001). The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29 4319–4333. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM-61148/GM/NIGMS NIH HHS/United States
- T32 CA009110/CA/NCI NIH HHS/United States
- T32 CA-09110/CA/NCI NIH HHS/United States
- R01 GM061148/GM/NIGMS NIH HHS/United States
- T32 ES007141/ES/NIEHS NIH HHS/United States
- T32 ES-07141/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases