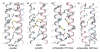Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino acid substitution - PubMed (original) (raw)
Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino acid substitution
Maneesh K Yadav et al. Biochemistry. 2006.
Abstract
A detailed understanding of the mechanisms by which particular amino acid sequences can give rise to more than one folded structure, such as for proteins that undergo large conformational changes or misfolding, is a long-standing objective of protein chemistry. Here, we describe the crystal structures of a single coiled-coil peptide in distinct parallel and antiparallel tetrameric configurations and further describe the parallel or antiparallel crystal structures of several related peptide sequences; the antiparallel tetrameric assemblies represent the first crystal structures of GCN4-derived peptides exhibiting such a configuration. Intriguingly, substitution of a single solvent-exposed residue enabled the parallel coiled-coil tetramer GCN4-pLI to populate the antiparallel configuration, suggesting that the two configurations are close enough in energy for subtle sequence changes to have important structural consequences. We present a structural analysis of the small changes to the helix register and side-chain conformations that accommodate the two configurations and have supplemented these results using solution studies and a molecular dynamics energetic analysis using a replica exchange methodology. Considering the previous examples of structural nonspecificity in coiled-coil peptides, the findings reported here not only emphasize the predisposition of the coiled-coil motif to adopt multiple configurations but also call attention to the associated risk that observed crytstal structures may not represent the only (or even the major) species present in solution.
Figures
Figure 1
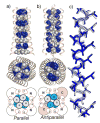
Crystal structures of the E20S variant (peptide 4) in the parallel and antiparallel tetrameric configurations. Side and top views show the parallel (a) and antiparallel (b) structures, highlighting core Leu (white) and Ile (blue) residues, with schematic diagrams showing interhelical packing interactions. c) Superposition of single helices from the antiparallel (blue) and parallel (beige) configurations. Backbone, β-carbons, and core residue heavy atoms are shown.
Figure 2
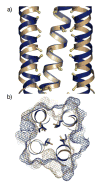
Comparison of hydrophobic packing interactions in the parallel and antiparallel E20S configurations. a) An overlay of the antiparallel (blue) and parallel (beige) E20S structures shows the shifted register for two helices in the antiparallel tetramer (with one helix omitted for clarity). Cα-Cβ bonds of hydrophobic core residues are shown, with Cβ atoms colored yellow, to illustrate the altered core geometry imposed by the antiparallel configuration. b) Cross-section of the same superposition showing the solvent-exposed surfaces of the antiparallel (blue mesh) and parallel (beige mesh) tetramers, along with side chain atoms for hydrophobic core residues. While the surfaces of the two assemblies are dissimilar, the core residues closely overlap.
Figure 3
Electrostatic interaction surfaces for the parallel and antiparallel coiled-coil tetramers. Dotted black lines are shown for potential polar intra- and interhelical interactions (< 4 Å) between surface residues for a) GCN4-pLI, b) E20S in the parallel configuration, c) E20S in the antiparallel configuration (K15 face), and d) E20S in the antiparallel configuration (S20 face). Positions 15 and 20 are colored yellow. Bridging water molecules are shown as red spheres.
Figure 4
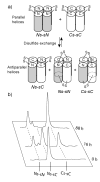
Equilibrium disulfide exchange assay for E20S (
). a) Assuming the glycyl linkers allow random sorting of the terminal thiols, tetramers in the parallel configuration will form only parallel homodimeric disulfides, whereas antiparallel tetramers will also form the antiparallel heterodimeric disulfide. b) HPLC traces showing disulfide exchange over the course of the equilibration. The disulfide-bonded antiparallel heterodimer (Ns-sC, 30 μM) rearranges to form only the parallel homodimers (Ns-sN and Cs-sC). In an analogous experiment initiated with equimolar amounts of the parallel dimers, no change was observed after 88 h, indicating the system is at equilibrium. Peptide 12 has a larger extinction coefficient than peptide 11 because it contains an added tyrosine residue to facilitate HPLC separation. The small peak eluting between Ns-sN and Ns-sC is a disulfide adduct of glutathione to peptide 11.
Similar articles
- A parallel coiled-coil tetramer with offset helices.
Liu J, Deng Y, Zheng Q, Cheng CS, Kallenbach NR, Lu M. Liu J, et al. Biochemistry. 2006 Dec 26;45(51):15224-31. doi: 10.1021/bi061914m. Epub 2006 Nov 29. Biochemistry. 2006. PMID: 17176044 - Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat.
Deng Y, Liu J, Zheng Q, Eliezer D, Kallenbach NR, Lu M. Deng Y, et al. Structure. 2006 Feb;14(2):247-55. doi: 10.1016/j.str.2005.10.010. Structure. 2006. PMID: 16472744 Free PMC article. - A seven-helix coiled coil.
Liu J, Zheng Q, Deng Y, Cheng CS, Kallenbach NR, Lu M. Liu J, et al. Proc Natl Acad Sci U S A. 2006 Oct 17;103(42):15457-62. doi: 10.1073/pnas.0604871103. Epub 2006 Oct 9. Proc Natl Acad Sci U S A. 2006. PMID: 17030805 Free PMC article. - Pharmacological interference with protein-protein interactions mediated by coiled-coil motifs.
Strauss HM, Keller S. Strauss HM, et al. Handb Exp Pharmacol. 2008;(186):461-82. doi: 10.1007/978-3-540-72843-6_19. Handb Exp Pharmacol. 2008. PMID: 18491064 Review. - The design of coiled-coil structures and assemblies.
Woolfson DN. Woolfson DN. Adv Protein Chem. 2005;70:79-112. doi: 10.1016/S0065-3233(05)70004-8. Adv Protein Chem. 2005. PMID: 15837514 Review.
Cited by
- The effects of pK(a) tuning on the thermodynamics and kinetics of folding: design of a solvent-shielded carboxylate pair at the a-position of a coiled-coil.
Lau WL, Degrado WF, Roder H. Lau WL, et al. Biophys J. 2010 Oct 6;99(7):2299-308. doi: 10.1016/j.bpj.2010.07.059. Biophys J. 2010. PMID: 20923665 Free PMC article. - Increasing the affinity of selective bZIP-binding peptides through surface residue redesign.
Kaplan JB, Reinke AW, Keating AE. Kaplan JB, et al. Protein Sci. 2014 Jul;23(7):940-53. doi: 10.1002/pro.2477. Epub 2014 Apr 30. Protein Sci. 2014. PMID: 24729132 Free PMC article. - Unique features of the anti-parallel, heterodimeric coiled-coil interaction between methyl-cytosine binding domain 2 (MBD2) homologues and GATA zinc finger domain containing 2A (GATAD2A/p66α).
Walavalkar NM, Gordon N, Williams DC Jr. Walavalkar NM, et al. J Biol Chem. 2013 Feb 1;288(5):3419-27. doi: 10.1074/jbc.M112.431346. Epub 2012 Dec 13. J Biol Chem. 2013. PMID: 23239876 Free PMC article. - The angiogenic inhibitor long pentraxin PTX3 forms an asymmetric octamer with two binding sites for FGF2.
Inforzato A, Baldock C, Jowitt TA, Holmes DF, Lindstedt R, Marcellini M, Rivieccio V, Briggs DC, Kadler KE, Verdoliva A, Bottazzi B, Mantovani A, Salvatori G, Day AJ. Inforzato A, et al. J Biol Chem. 2010 Jun 4;285(23):17681-92. doi: 10.1074/jbc.M109.085639. Epub 2010 Apr 2. J Biol Chem. 2010. PMID: 20363749 Free PMC article. - Computational analysis of residue contributions to coiled-coil topology.
Ramos J, Lazaridis T. Ramos J, et al. Protein Sci. 2011 Nov;20(11):1845-55. doi: 10.1002/pro.718. Epub 2011 Sep 20. Protein Sci. 2011. PMID: 21858887 Free PMC article.
References
- Crick FHC. The packing of alpha-helices: simple coiled-coils. Acta Cryst. 1953;6:689–97.
- Alber T. How GCN4 binds DNA. Curr Biol. 1993;3:182–4. - PubMed
- O’Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–44. - PubMed
- Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–7. - PubMed
- Harbury PB, Kim PS, Alber T. Crystal structure of an isoleucine-zipper trimer. Nature. 1994;371:80–3. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous