Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica - PubMed (original) (raw)
Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica
Joseph T Penrod et al. J Bacteriol. 2006 Apr.
Abstract
Salmonellae can use ethanolamine (EA) as a sole source of carbon and nitrogen. This ability is encoded by an operon (eut) containing 17 genes, only 6 of which are required under standard conditions (37 degrees C; pH 7.0). Five of the extra genes (eutM, -N, -L, -K, and -G) become necessary under conditions that favor loss of the volatile intermediate, acetaldehyde, which escapes as a gas during growth on EA and is lost at a higher rate from these mutants. The eutM, -N, -L, and -K genes encode homologues of shell proteins of the carboxysome, an organelle shown (in other organisms) to concentrate CO(2). We propose that carboxysome-like organelles help bacteria conserve certain volatile metabolites-CO(2) or acetaldehyde-perhaps by providing a low-pH compartment. The EutG enzyme converts acetaldehyde to ethanol, which may improve carbon retention by forming acetals; alternatively, EutG may recycle NADH within the carboxysome.
Figures
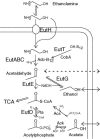
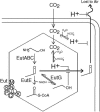
FIG. 1.
Basic ethanolamine metabolism. EA can enter the cell by diffusion, either via EutH or by other, unidentified routes. The diagram depicts the known roles of eut enzymes in metabolism of EA. The loss of acetaldehyde to the gas phase proposed here is indicated by a dotted arrow.
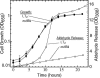
FIG. 2.
Growth and aldehyde release by wild-type cells. Growth (closed symbols) and aldehyde release (open symbols) of strain LT2 grown on minimal acetate (squares), on glycerol (triangles, points up), or on glycerol plus ethanolamine and B12 (triangles, points down). Minimal medium was NCE salts (pH 7.0), which provides ammonia as a nitrogen source. Cultures were grown at 37°C, and ethanolamine was provided at 30 mM. The error bars represent standard deviations.
FIG. 3.
Aldehyde release requires ethanolamine ammonia lyase. Growth (closed symbols) and aldehyde release (open symbols) of LT2 (squares) and a eutB mutant (circles) on minimal glycerol, ethanolamine, and B12 medium. Ethanolamine was provided at 15 mM. The error bars represent standard deviations.
FIG. 4.
Mutations that block acetaldehyde metabolism increase aldehyde release. Mutants eutB (circles), eutD (diamonds), eutE (triangles, points up), and eutG (triangles, points down) were compared to wild-type LT2 (squares) for aldehyde release during growth on glycerol, ethanolamine, and B12. Ethanolamine was provided at 15 mM.
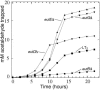
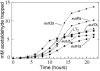
FIG. 5.
Aldehyde release increases in carboxysome mutants. Mutants eutN (circles), eutM (diamonds), eutL (triangles, points up), eutK (right triangles), and eutS (triangles, points down) were compared to wild-type LT2 (squares) for aldehyde release during growth on glycerol, ethanolamine, and B12. Ethanolamine was provided at 15 mM.
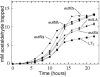
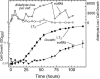
FIG. 6.
Effects of other eut mutations on aldehyde release. Mutants eutH (circles), eutJ (diamonds), eutQ (triangles, points up), eutT (right triangles), and eutP (triangles, points down) were compared to wild-type LT2 (squares) for aldehyde release during growth on glycerol, ethanolamine, and B12. Ethanolamine was provided at 15 mM.
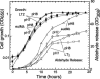
FIG. 7.
Aldehyde release during growth on ethanolamine as a carbon source; growth (closed symbols) and aggregated aldehyde release normalized to cell growth (open symbols). Wild-type LT2 (squares) was compared to a eutM deletion mutant (triangles, points down). Ethanolamine (40 mM) was the sole carbon source. The error bars represent standard deviations.
FIG. 8.
Effect of pH on aldehyde release. Growth (closed symbols) and cumulative aldehyde release (open symbols) were measured at pH 7 and 8. Wild-type LT2 is depicted by squares at pH 7 and diamonds at pH 8. A eutM deletion mutant is depicted at pH 7 (triangles, points down) and pH 8 (triangles, points up). The medium was buffered with 50 mM MOPS (pH 7) or Bicine (pH 8) and contained glycerol, ethanolamine (20 mM), and B12. The error bars represent standard deviations.
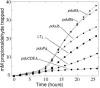
FIG. 9.
Aldehyde release by pdu mutants. Mutants pduCDE (circles), pduP (triangles, points down), pduB (diamonds), pduJ (triangles, points up), and pduK (right triangles) were compared to wild-type LT2 (squares) during growth on pyruvate (40 mM), 1,2-propanediol (80 mM), and B12. The cultures were buffered at 7.0 with NCE salts and incubated at 38°C. All strains grew at the same rate (data not shown).
FIG. 10.
Proposed role of the carboxysome. It is proposed here that the primary function of a carboxysome is to retain and prevent loss of a volatile metabolite, acetaldehyde in the case of the ethanolamine pathway. Retention of volatile aldehydes (in Salmonella) and retention of CO2 (in other organisms) might be explained if the carboxysome provided a low-pH compartment.
Similar articles
- Minimal functions and physiological conditions required for growth of salmonella enterica on ethanolamine in the absence of the metabolosome.
Brinsmade SR, Paldon T, Escalante-Semerena JC. Brinsmade SR, et al. J Bacteriol. 2005 Dec;187(23):8039-46. doi: 10.1128/JB.187.23.8039-8046.2005. J Bacteriol. 2005. PMID: 16291677 Free PMC article. - Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster.
Stojiljkovic I, Bäumler AJ, Heffron F. Stojiljkovic I, et al. J Bacteriol. 1995 Mar;177(5):1357-66. doi: 10.1128/jb.177.5.1357-1366.1995. J Bacteriol. 1995. PMID: 7868611 Free PMC article. - The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins.
Kofoid E, Rappleye C, Stojiljkovic I, Roth J. Kofoid E, et al. J Bacteriol. 1999 Sep;181(17):5317-29. doi: 10.1128/JB.181.17.5317-5329.1999. J Bacteriol. 1999. PMID: 10464203 Free PMC article. - Self-assembly in the carboxysome: a viral capsid-like protein shell in bacterial cells.
Yeates TO, Tsai Y, Tanaka S, Sawaya MR, Kerfeld CA. Yeates TO, et al. Biochem Soc Trans. 2007 Jun;35(Pt 3):508-11. doi: 10.1042/BST0350508. Biochem Soc Trans. 2007. PMID: 17511640 Review. - Ethanolamine Utilization in Bacteria.
Kaval KG, Garsin DA. Kaval KG, et al. mBio. 2018 Feb 20;9(1):e00066-18. doi: 10.1128/mBio.00066-18. mBio. 2018. PMID: 29463652 Free PMC article. Review.
Cited by
- Engineered protein nano-compartments for targeted enzyme localization.
Choudhary S, Quin MB, Sanders MA, Johnson ET, Schmidt-Dannert C. Choudhary S, et al. PLoS One. 2012;7(3):e33342. doi: 10.1371/journal.pone.0033342. Epub 2012 Mar 12. PLoS One. 2012. PMID: 22428024 Free PMC article. - Two-dimensional crystals of carboxysome shell proteins recapitulate the hexagonal packing of three-dimensional crystals.
Dryden KA, Crowley CS, Tanaka S, Yeates TO, Yeager M. Dryden KA, et al. Protein Sci. 2009 Dec;18(12):2629-35. doi: 10.1002/pro.272. Protein Sci. 2009. PMID: 19844993 Free PMC article. - Microbial byproducts determine reproductive fitness of free-living and parasitic nematodes.
Venzon M, Das R, Luciano DJ, Burnett J, Park HS, Devlin JC, Kool ET, Belasco JG, Hubbard EJA, Cadwell K. Venzon M, et al. Cell Host Microbe. 2022 Jun 8;30(6):786-797.e8. doi: 10.1016/j.chom.2022.03.015. Epub 2022 Apr 11. Cell Host Microbe. 2022. PMID: 35413267 Free PMC article. - Preliminary structural investigations of the Eut-L shell protein of the ethanolamine ammonia-lyase metabolosome of Escherichia coli.
Nikolakakis K, Ohtaki A, Newton K, Chworos A, Sagermann M. Nikolakakis K, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009 Feb 1;65(Pt 2):128-32. doi: 10.1107/S1744309108042127. Epub 2009 Jan 31. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009. PMID: 19194002 Free PMC article. - The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica.
Cheng S, Fan C, Sinha S, Bobik TA. Cheng S, et al. PLoS One. 2012;7(10):e47144. doi: 10.1371/journal.pone.0047144. Epub 2012 Oct 15. PLoS One. 2012. PMID: 23077559 Free PMC article.
References
- Badger, M. R., and G. D. Price. 2003. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 54:609-622. - PubMed
- Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967-5975. - PMC - PubMed
- Bradbeer, C. 1965. The clostridial fermentations of choline and ethanolamine. I. Preparation and properties of cell-free extracts. J. Biol. Chem. 240:4669-4674. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases