Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody - PubMed (original) (raw)
Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody
Ponraj Prabakaran et al. J Biol Chem. 2006.
Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV, or SCV), which caused a world-wide epidemic in 2002 and 2003, binds to a receptor, angiotensin-converting enzyme 2 (ACE2), through the receptor-binding domain (RBD) of its envelope (spike, S) glycoprotein. The RBD is very immunogenic; it is a major SCV neutralization determinant and can elicit potent neutralizing antibodies capable of out-competing ACE2. However, the structural basis of RBD immunogenicity, RBD-mediated neutralization, and the role of RBD in entry steps following its binding to ACE2 have not been elucidated. By mimicking immune responses with the use of RBD as an antigen to screen a large human antibody library derived from healthy volunteers, we identified a novel potent cross-reactive SCV-neutralizing monoclonal antibody, m396, which competes with ACE2 for binding to RBD, and determined the crystal structure of the RBD-antibody complex at 2.3-A resolution. The antibody-bound RBD structure is completely defined, revealing two previously unresolved segments (residues 376-381 and 503-512) and a new disulfide bond (between residues 378 and 511). Interestingly, the overall structure of the m396-bound RBD is not significantly different from that of the ACE2-bound RBD. The antibody epitope is dominated by a 10-residue-long protruding beta6-beta7 loop with two putative ACE2-binding hotspot residues (Ile-489 and Tyr-491). These results provide a structural rationale for the function of a major determinant of SCV immunogenicity and neutralization, the development of SCV therapeutics based on the antibody paratope and epitope, and a retrovaccinology approach for the design of anti-SCV vaccines. The available structural information indicates that the SCV entry may not be mediated by ACE2-induced conformational changes in the RBD but may involve other conformational changes or/and yet to be identified coreceptors.
Figures
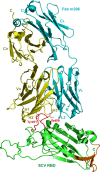
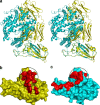
FIGURE 1
Overall structure of the SCV RBD in complex with the neutralizing antibody Fab m396. The SCV RBD is in green, and the prominent neutralizing site comprising residues 482 through 491 (β6–β7 loop) is in red. The side chains of two important residues, Ile-489 and Tyr-491, of the loop are shown as sticks. A portion of the structure shown in brown constitutes the 16 amino acid residues that were not observed in the RBD·ACE2 structure (20). The light and heavy chains of the Fab are shown in cyan and yellow, respectively, with labeled CDRs, H1, H2, H3, and L3, which make contacts with the RBD.
FIGURE 2
Comparison of the RBD·Fab and the RBD·ACE2 structures. The newly identified two segments of RBD in the Fab complex are denoted by blue and pink colors. a, sequence and secondary structure assignment of RBD. b, structural alignment between RBD structures based on Cα positions. RBD structures from the Fab and ACE2 complexes are shown in green and cyan, respectively. Blue and pink segments defined in the RBD-Fab structure revealed the fourth disulfide bond within the RBD (between residues Cys-378 and Cys-511), which is shown as a stick model. c and d, stereoviews showing the 2_Fo_ – Fc electron density maps contoured at 1.0 σ level along the segments 375–382 and 501–512.
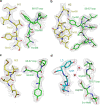
FIGURE 3
Critical interactions between SCV RBD (green) and Fab m396 (yellow and cyan for heavy- and light-chain CDRs, respectively) depicted with 2_Fo_ – Fc electron density maps contoured at the 1.0 σ level. CDRs H1, H2, and H3 recognize the major neutralizing site, the β6–β7 loop. L3 exclusively contacts minor binding sites with the involvement of bridging water molecules. a, H1 residues Ser-31 and Thr-33 form hydrogen bonds with RBD residues Thr-486 and Thr-488 via backbone-side chain interactions. b, H2 displays a concave surface and contributes to the specific interactions between H2 residues Thr-52 and Asn-58, and RBD residue Tyr-491. c, H3 residue Val-97 contacts the RBD and buries the largest surface area per residue (108 Ä2) among all residues of the antibody combining site. The carbonyl of Val-97 forms a hydrogen bond to the side-chain amide of Gln-492 of RBD. d, L3 is the only light chain CDR that binds to the RBD with two bridging water molecules (pink spheres) involved.
FIGURE 4
Structural features facilitating the binding between the SCV RBD and the Fab m396 include shape complementarity and specific side-chain interactions.a, the protruding structure of RBD (highlighted in yellow) within 3.5 Ä of the antibody surface approximates the antibody epitope on the RBD (green). b, the RBD-interacting area of the antibody (highlighted in green) within 3.5 Ä of the RBD surface approximates the paratope of the antibody (yellow), showing high structural complementarity to the antibody epitope shown in yellow in panel a.
FIGURE 5
Stereoview of superimposed structures of RBD-free (red) and RBD-bound Fab m396 (blue) based on the Cα positions in their variable domains, which shows a significant elbow-angle difference between the two conformations of Fab m396. The unliganded Fab has an open or straight elbow angle, whereas the SCV RBD-bound Fab has a closed or highly bent elbow angle.
FIGURE 6
Comparison of the RBD·Fab and the RBD·ACE2 complexes: lack of induced conformational changes of RBD and overlapping but not identical binding sites of Fab and ACE2.a, stereoview of the superposition of the RBD-antibody (in yellow) and RBD-receptor (in cyan) complexes. The superposition was based on the alignment of the Cα positions of RBD in the two complexes. The heavy chain of the antibody significantly overlaps with the receptor. A common binding region, the β6–β7 loop of RBD, is shared by the antibody and the receptor. The residues in the common binding region of RBD may contribute to the orientation of the antibody and the receptor to the spike on viral surface, the high affinity of the antibody binding, and the ACE2 binding specificities that lead to SARS infection of humans and cross-species transmission. b, structural footprints of the antibody and c, the receptor on the SCV RBD shown as red batches on the RBD surface. The circled area on the RBD surface represents the common binding site for the antibody and the receptor.
Similar articles
- Effects of human anti-spike protein receptor binding domain antibodies on severe acute respiratory syndrome coronavirus neutralization escape and fitness.
Sui J, Deming M, Rockx B, Liddington RC, Zhu QK, Baric RS, Marasco WA. Sui J, et al. J Virol. 2014 Dec;88(23):13769-80. doi: 10.1128/JVI.02232-14. Epub 2014 Sep 17. J Virol. 2014. PMID: 25231316 Free PMC article. - Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R.
Hwang WC, Lin Y, Santelli E, Sui J, Jaroszewski L, Stec B, Farzan M, Marasco WA, Liddington RC. Hwang WC, et al. J Biol Chem. 2006 Nov 10;281(45):34610-6. doi: 10.1074/jbc.M603275200. Epub 2006 Sep 5. J Biol Chem. 2006. PMID: 16954221 Free PMC article. - Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein.
He Y, Li J, Li W, Lustigman S, Farzan M, Jiang S. He Y, et al. J Immunol. 2006 May 15;176(10):6085-92. doi: 10.4049/jimmunol.176.10.6085. J Immunol. 2006. PMID: 16670317 - Structural Basis of a Human Neutralizing Antibody Specific to the SARS-CoV-2 Spike Protein Receptor-Binding Domain.
Yang M, Li J, Huang Z, Li H, Wang Y, Wang X, Kang S, Huang X, Wu C, Liu T, Jia Z, Liang J, Yuan X, He S, Chen X, Zhou Z, Chen Q, Liu S, Li J, Zheng H, Liu X, Li K, Yao X, Lang B, Liu L, Liao HX, Chen S. Yang M, et al. Microbiol Spectr. 2021 Oct 31;9(2):e0135221. doi: 10.1128/Spectrum.01352-21. Epub 2021 Oct 13. Microbiol Spectr. 2021. PMID: 34643438 Free PMC article. - Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions.
Li W, Wong SK, Li F, Kuhn JH, Huang IC, Choe H, Farzan M. Li W, et al. J Virol. 2006 May;80(9):4211-9. doi: 10.1128/JVI.80.9.4211-4219.2006. J Virol. 2006. PMID: 16611880 Free PMC article. Review. No abstract available.
Cited by
- A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Kreye J, Reincke SM, Kornau HC, Sánchez-Sendin E, Corman VM, Liu H, Yuan M, Wu NC, Zhu X, Lee CD, Trimpert J, Höltje M, Dietert K, Stöffler L, von Wardenburg N, van Hoof S, Homeyer MA, Hoffmann J, Abdelgawad A, Gruber AD, Bertzbach LD, Vladimirova D, Li LY, Barthel PC, Skriner K, Hocke AC, Hippenstiel S, Witzenrath M, Suttorp N, Kurth F, Franke C, Endres M, Schmitz D, Jeworowski LM, Richter A, Schmidt ML, Schwarz T, Müller MA, Drosten C, Wendisch D, Sander LE, Osterrieder N, Wilson IA, Prüss H. Kreye J, et al. Cell. 2020 Nov 12;183(4):1058-1069.e19. doi: 10.1016/j.cell.2020.09.049. Epub 2020 Sep 23. Cell. 2020. PMID: 33058755 Free PMC article. - The landscape of potential health benefits of carotenoids as natural supportive therapeutics in protecting against Coronavirus infection.
Lu LW, Gao Y, Quek SY, Foster M, Eason CT, Liu M, Wang M, Chen JH, Chen F. Lu LW, et al. Biomed Pharmacother. 2022 Oct;154:113625. doi: 10.1016/j.biopha.2022.113625. Epub 2022 Aug 31. Biomed Pharmacother. 2022. PMID: 36058151 Free PMC article. Review. - Analysis of the molecular mechanism of SARS-CoV-2 antibodies.
Jin D, Wei J, Sun J. Jin D, et al. Biochem Biophys Res Commun. 2021 Aug 20;566:45-52. doi: 10.1016/j.bbrc.2021.06.001. Epub 2021 Jun 5. Biochem Biophys Res Commun. 2021. PMID: 34116356 Free PMC article. Review. - Removal of a C-terminal serine residue proximal to the inter-chain disulfide bond of a human IgG1 lambda light chain mediates enhanced antibody stability and antibody dependent cell-mediated cytotoxicity.
Shen Y, Zeng L, Zhu A, Blanc T, Patel D, Pennello A, Bari A, Ng S, Persaud K, Kang YK, Balderes P, Surguladze D, Hindi S, Zhou Q, Ludwig DL, Snavely M. Shen Y, et al. MAbs. 2013 May-Jun;5(3):418-31. doi: 10.4161/mabs.24291. Epub 2013 Apr 8. MAbs. 2013. PMID: 23567210 Free PMC article. - Computational design and modeling of nanobodies toward SARS-CoV-2 receptor binding domain.
Yang J, Zhang Z, Yang F, Zhang H, Wu H, Zhu F, Xue W. Yang J, et al. Chem Biol Drug Des. 2021 Jul;98(1):1-18. doi: 10.1111/cbdd.13847. Epub 2021 May 13. Chem Biol Drug Des. 2021. PMID: 33894099 Free PMC article.
References
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
- Drosten C., Gunther S., Preiser W., van der W.S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
- Holmes K.V. N. Engl. J. Med. 2003;348:1948–1951. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous