Identification and quantification of archaea involved in primary endodontic infections - PubMed (original) (raw)
Identification and quantification of archaea involved in primary endodontic infections
M E Vianna et al. J Clin Microbiol. 2006 Apr.
Abstract
Members of the domain Archaea, one of the three domains of life, are a highly diverse group of prokaryotes, distinct from bacteria and eukaryotes. Despite their abundance and ubiquity on earth, including their close association with humans, animals, and plants, so far no pathogenic archaea have been described. As some archaea live in close proximity to anaerobic bacteria, for instance, in the human gut system and in periodontal pockets, the aim of our study was to assess whether archaea might possibly be involved in human endodontic infections, which are commonly polymicrobial. We analyzed 20 necrotic uniradicular teeth with radiographic evidence of apical periodontitis and with no previous endodontic treatment. Using real-time quantitative PCR based on the functional gene mcrA (encoding the methyl coenzyme M reductase, specific to methanogenic archaea) and on archaeal 16S rRNA genes, we found five cases to be positive. Direct sequencing of PCR products from both genes showed that the archaeal community was dominated by a Methanobrevibacter oralis-like phylotype. The size of the archaeal population at the diseased sites ranged from 1.3 x 10(5) to 6.8 x 10(5) 16S rRNA gene target molecule numbers and accounted for up to 2.5% of the total prokaryotic community (i.e., bacteria plus archaea). Our findings show that archaea can be intimately connected with infectious diseases and thus support the hypothesis that members of the domain Archaea may have a role as human pathogens.
Figures
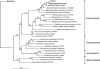
FIG. 1.
Phylogenetic tree showing the positions of 16S rRNA gene types identified in infected root canals of human teeth relative to those of representative members of the four major lineages from the domain Archaea. Sequences determined in this study are shown in boldface. The scale bar corresponds to 0.1 substitution per nucleotide. The tree was calculated using 798 nucleotide positions and the neighbor-joining approach (with the Felsenstein correction), via the ARB program package (23). The statistical significance levels of interior nodes, shown as percentages, were determined by performing bootstrap analyses (1,000 replications; only values over 50% are shown).
FIG. 2.
Phylogenetic tree showing the positions of the mcrA gene types identified in infected root canals of human teeth relative to those of representative members of methanogenic archaea. Sequences determined in this study are shown in boldface. The scale bar corresponds to 0.1 substitution per nucleotide. The tree was calculated using 464 nucleotide positions and the neighbor-joining approach (with the Felsenstein correction), via the ARB program package (23). The statistical significance levels of interior nodes, shown as percentages, were determined by performing bootstrap analyses (1,000 replications; only values over 50% are shown).
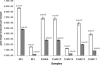
FIG. 3.
Comparison of 16S rRNA gene target molecule numbers (white bars) with mcrA gene target molecule numbers (gray bars), determined by RTQ-PCR from pure cultures and from samples obtained from primary endodontic infections. Error bars indicate standard deviations from three replicate RTQ-PCR runs. Mo, M. oralis; Ms, M. smithii.
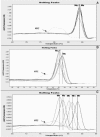
FIG. 4.
Dissociation curves of archaeal 16S rRNA gene PCR products (A) and methanogenic mcrA gene PCR products (B and C). E, endodontic samples; Mo, M. oralis; Ms, M. smithii; Mb, M. bryantii; Mm, M. maripaludis; Mh, M. hungatei; NTC, nontarget control.
Similar articles
- T-RFLP-based mcrA gene analysis of methanogenic archaea in association with oral infections and evidence of a novel Methanobrevibacter phylotype.
Vianna ME, Conrads G, Gomes BP, Horz HP. Vianna ME, et al. Oral Microbiol Immunol. 2009 Oct;24(5):417-22. doi: 10.1111/j.1399-302X.2009.00539.x. Oral Microbiol Immunol. 2009. PMID: 19702957 - Characterization of C1-metabolizing prokaryotic communities in methane seep habitats at the Kuroshima Knoll, southern Ryukyu Arc, by analyzing pmoA, mmoX, mxaF, mcrA, and 16S rRNA genes.
Inagaki F, Tsunogai U, Suzuki M, Kosaka A, Machiyama H, Takai K, Nunoura T, Nealson KH, Horikoshi K. Inagaki F, et al. Appl Environ Microbiol. 2004 Dec;70(12):7445-55. doi: 10.1128/AEM.70.12.7445-7455.2004. Appl Environ Microbiol. 2004. PMID: 15574947 Free PMC article. - Distribution of Archaea in Japanese patients with periodontitis and humoral immune response to the components.
Yamabe K, Maeda H, Kokeguchi S, Tanimoto I, Sonoi N, Asakawa S, Takashiba S. Yamabe K, et al. FEMS Microbiol Lett. 2008 Oct;287(1):69-75. doi: 10.1111/j.1574-6968.2008.01304.x. Epub 2008 Aug 14. FEMS Microbiol Lett. 2008. PMID: 18707623 - Methanogenic archaea in subgingival sites: a review.
Nguyen-Hieu T, Khelaifia S, Aboudharam G, Drancourt M. Nguyen-Hieu T, et al. APMIS. 2013 Jun;121(6):467-77. doi: 10.1111/apm.12015. Epub 2012 Oct 19. APMIS. 2013. PMID: 23078250 Review. - Meta-analyses on the Periodontal Archaeome.
de Cena JA, Silvestre-Barbosa Y, Belmok A, Stefani CM, Kyaw CM, Damé-Teixeira N. de Cena JA, et al. Adv Exp Med Biol. 2022;1373:69-93. doi: 10.1007/978-3-030-96881-6_4. Adv Exp Med Biol. 2022. PMID: 35612793 Review.
Cited by
- Methyl coenzyme M reductase (mcrA) gene abundance correlates with activity measurements of methanogenic H₂ /CO₂ -enriched anaerobic biomass.
Morris R, Schauer-Gimenez A, Bhattad U, Kearney C, Struble CA, Zitomer D, Maki JS. Morris R, et al. Microb Biotechnol. 2014 Jan;7(1):77-84. doi: 10.1111/1751-7915.12094. Epub 2013 Oct 31. Microb Biotechnol. 2014. PMID: 24320083 Free PMC article. - Oral Microbiome and Dental Caries Development.
Zhang JS, Chu CH, Yu OY. Zhang JS, et al. Dent J (Basel). 2022 Sep 30;10(10):184. doi: 10.3390/dj10100184. Dent J (Basel). 2022. PMID: 36285994 Free PMC article. Review. - First Insights into the Diverse Human Archaeome: Specific Detection of Archaea in the Gastrointestinal Tract, Lung, and Nose and on Skin.
Koskinen K, Pausan MR, Perras AK, Beck M, Bang C, Mora M, Schilhabel A, Schmitz R, Moissl-Eichinger C. Koskinen K, et al. mBio. 2017 Nov 14;8(6):e00824-17. doi: 10.1128/mBio.00824-17. mBio. 2017. PMID: 29138298 Free PMC article. - Archaea prevalence in inflamed pulp tissues.
Efenberger M, Agier J, Pawłowska E, Brzezińska-Błaszczyk E. Efenberger M, et al. Cent Eur J Immunol. 2015;40(2):194-200. doi: 10.5114/ceji.2015.51358. Epub 2015 Aug 3. Cent Eur J Immunol. 2015. PMID: 26557034 Free PMC article. - Endodontic microbiology.
Narayanan LL, Vaishnavi C. Narayanan LL, et al. J Conserv Dent. 2010 Oct;13(4):233-9. doi: 10.4103/0972-0707.73386. J Conserv Dent. 2010. PMID: 21217951 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials