Transmission of an uncultivated Bergeyella strain from the oral cavity to amniotic fluid in a case of preterm birth - PubMed (original) (raw)
Transmission of an uncultivated Bergeyella strain from the oral cavity to amniotic fluid in a case of preterm birth
Yiping W Han et al. J Clin Microbiol. 2006 Apr.
Abstract
Intrauterine infection is a recognized cause of preterm birth. The infectious organisms are believed to originate primarily from the vaginal tract and secondarily from other parts of the body. It is plausible that microbes in the oral cavity can be transmitted to the pregnant uterus. However, direct evidence supporting such a transmission is lacking. In this study, amniotic fluids of 34 pregnant women were examined by PCR using 16S and 23S rRNA universally conserved primers. Bacterial DNA was amplified from the only patient with clinical intrauterine infection and histologic necrotizing acute and chronic chorioamnionitis. One strain, Bergeyella sp. clone AF14, was detected and was 99.7% identical to a previously reported uncultivated oral Bergeyella strain, clone AK152, at the 16S rRNA level. The same strain was detected in the subgingival plaque of the patient but not in her vaginal tract. The 16S-23S rRNA sequence of clone AF14 matched exactly with the sequences amplified from the patient's subgingival plaque. These observations suggest that the Bergeyella strain identified in the patient's intrauterine infection originated from the oral cavity. This is the first direct evidence of oral-utero microbial transmission. The patient's periodontal health during pregnancy was unclear. She did not have detectable periodontal disease during postpartum examination. Bergeyella spp. had not been previously associated with preterm birth and were detected in subgingival plaque of women without clinical levels of intrauterine infection. Uncultivated species may be overlooked opportunistic pathogens in preterm birth. This study sheds new light on the implication of oral bacteria in preterm birth.
Figures
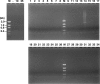
FIG. 1.
PCR examination of AF samples with universal primers 785F and 442R. The numbers at the top correspond to patient numbers. Lane M, DNA molecular size markers, as indicated on the left; Lane —, negative control.
FIG. 2.
I. DNA sequence comparison of 16S-23S rRNA genes of Bergeyella sp. clone AK152 (BergAK152), deposited by Paster et al.; clone AF14 (BergAF14), isolated from AF of patient 14; and clone P14 (BergP14), isolated from the subgingival plaque of patient 14. Identical nucleotides are highlighted in gray. Primers for PCR are indicated above or below the sequences. The dashed lines indicate that no sequence is available. II. Schematic diagram of 16S-23S rRNA genes. Primer locations and their corresponding products are indicated by arrows and lines.
FIG. 2.
I. DNA sequence comparison of 16S-23S rRNA genes of Bergeyella sp. clone AK152 (BergAK152), deposited by Paster et al.; clone AF14 (BergAF14), isolated from AF of patient 14; and clone P14 (BergP14), isolated from the subgingival plaque of patient 14. Identical nucleotides are highlighted in gray. Primers for PCR are indicated above or below the sequences. The dashed lines indicate that no sequence is available. II. Schematic diagram of 16S-23S rRNA genes. Primer locations and their corresponding products are indicated by arrows and lines.
FIG. 2.
I. DNA sequence comparison of 16S-23S rRNA genes of Bergeyella sp. clone AK152 (BergAK152), deposited by Paster et al.; clone AF14 (BergAF14), isolated from AF of patient 14; and clone P14 (BergP14), isolated from the subgingival plaque of patient 14. Identical nucleotides are highlighted in gray. Primers for PCR are indicated above or below the sequences. The dashed lines indicate that no sequence is available. II. Schematic diagram of 16S-23S rRNA genes. Primer locations and their corresponding products are indicated by arrows and lines.
FIG. 3.
Direct and nested PCR analyses of samples collected from patient 14. a. PCR using universal primers 785F and 442R. The products obtained were used as temples for nested PCR in panels b to e. b. Nested PCR using universal primers 785F and L189R. c. Nested PCR using _Bergeyella_-specific primer BergF and universal primer L189R. d. Nested PCR using _Bergeyella_-specific primers BergF and 14BitsR1. e. Nested PCR using _Bergeyella_-specific primers BergF and 14BitsR2. Lanes M, molecular size markers, as indicated on the left; lanes A, AF; lanes P, subgingival plaque; lanes V, vaginal swab.
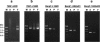
FIG. 4.
Detection of Bergeyella in subgingival plaque samples by nested PCR using primers BergF and 14BitsR1. The numbers on the top correspond to the patient numbers. Lane M, DNA molecular size markers, as indicated on the left; Lane —, negative control.
Similar articles
- Periodontitis, a marker of risk in pregnancy for preterm birth.
Dörtbudak O, Eberhardt R, Ulm M, Persson GR. Dörtbudak O, et al. J Clin Periodontol. 2005 Jan;32(1):45-52. doi: 10.1111/j.1600-051X.2004.00630.x. J Clin Periodontol. 2005. PMID: 15642058 - Detection of Porphyromonas gingivalis in the amniotic fluid in pregnant women with a diagnosis of threatened premature labor.
León R, Silva N, Ovalle A, Chaparro A, Ahumada A, Gajardo M, Martinez M, Gamonal J. León R, et al. J Periodontol. 2007 Jul;78(7):1249-55. doi: 10.1902/jop.2007.060368. J Periodontol. 2007. PMID: 17608580 - Preterm birth: associations with genital and possibly oral microflora.
Hill GB. Hill GB. Ann Periodontol. 1998 Jul;3(1):222-32. doi: 10.1902/annals.1998.3.1.222. Ann Periodontol. 1998. PMID: 9722706 Review. - Can oral anaerobic bacteria cause adverse pregnancy outcomes?
Andonova I, Iliev V, Živković N, Sušič E, Bego I, Kotevska V. Andonova I, et al. Pril (Makedon Akad Nauk Umet Odd Med Nauki). 2015;36(1):137-43. Pril (Makedon Akad Nauk Umet Odd Med Nauki). 2015. PMID: 26076783 - Periodontal disease and preterm birth relationship: a review of the literature.
Sacco G, Carmagnola D, Abati S, Luglio PF, Ottolenghi L, Villa A, Maida C, Campus G. Sacco G, et al. Minerva Stomatol. 2008 May;57(5):233-46, 246-50. Minerva Stomatol. 2008. PMID: 18496486 Review. English, Italian.
Cited by
- Individualized medicine and the microbiome in reproductive tract.
Braundmeier AG, Lenz KM, Inman KS, Chia N, Jeraldo P, Walther-António MR, Berg Miller ME, Yang F, Creedon DJ, Nelson H, White BA. Braundmeier AG, et al. Front Physiol. 2015 Apr 1;6:97. doi: 10.3389/fphys.2015.00097. eCollection 2015. Front Physiol. 2015. PMID: 25883569 Free PMC article. Review. - Role of Maternal Infections and Inflammatory Responses on Craniofacial Development.
Bhagirath AY, Medapati MR, de Jesus VC, Yadav S, Hinton M, Dakshinamurti S, Atukorallaya D. Bhagirath AY, et al. Front Oral Health. 2021 Sep 6;2:735634. doi: 10.3389/froh.2021.735634. eCollection 2021. Front Oral Health. 2021. PMID: 35048051 Free PMC article. Review. - Bacterial aetiological agents of intra-amniotic infections and preterm birth in pregnant women.
Mendz GL, Kaakoush NO, Quinlivan JA. Mendz GL, et al. Front Cell Infect Microbiol. 2013 Oct 16;3:58. doi: 10.3389/fcimb.2013.00058. eCollection 2013. Front Cell Infect Microbiol. 2013. PMID: 24137568 Free PMC article. Review. - Term stillbirth caused by oral Fusobacterium nucleatum.
Han YW, Fardini Y, Chen C, Iacampo KG, Peraino VA, Shamonki JM, Redline RW. Han YW, et al. Obstet Gynecol. 2010 Feb;115(2 Pt 2):442-445. doi: 10.1097/AOG.0b013e3181cb9955. Obstet Gynecol. 2010. PMID: 20093874 Free PMC article. - Mobile microbiome: oral bacteria in extra-oral infections and inflammation.
Han YW, Wang X. Han YW, et al. J Dent Res. 2013 Jun;92(6):485-91. doi: 10.1177/0022034513487559. Epub 2013 Apr 26. J Dent Res. 2013. PMID: 23625375 Free PMC article. Review.
References
- Akcali, K. C., G. Gibori, and S. A. Khan. 2003. The involvement of apoptotic regulators during in vitro decidualization. Eur. J. Endocrinol. 149:69-75. - PubMed
- Alanen, A., and E. Laurikainen. 1999. Second-trimester abortion caused by Capnocytophaga sputigena: case report. Am. J. Perinatol. 16:181-183. - PubMed
- Altshuler, G., and S. Hyde. 1985. Fusobacteria. An important cause of chorioamnionitis. Arch. Pathol. Lab. Med. 109:739-743. - PubMed
- Andrews, W. W., R. L. Goldenberg, and J. C. Hauth. 1995. Preterm labor: emerging role of genital tract infections. Infect. Agents Dis. 4:196-211. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous