Introducing TreeClimber, a test to compare microbial community structures - PubMed (original) (raw)
Comparative Study
Introducing TreeClimber, a test to compare microbial community structures
Patrick D Schloss et al. Appl Environ Microbiol. 2006 Apr.
Abstract
The phylogenetic and ecological complexity of microbial communities necessitates the development of new methods to determine whether two or more communities have the same structure even though it is not possible to sample the communities exhaustively. To address this need, we adapted a method used in population genetics, the parsimony test, to determine the relatedness of communities. Here we describe our implementation of the parsimony test, TreeClimber, in which we reanalyzed six previously published studies and compared the results of the analysis to those obtained using integral-LIBSHUFF.
Figures
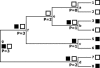
FIG. 1.
Examples of using Fitch's algorithm (11) to score a phylogenetic tree when samples are obtained from two communities. The letters represent treatments for the nodes, and the numbers represent the treatment label for each sequence considered. The parsimony scores (P) and treatment labels for each node as they aggregate through the nodes are indicated.
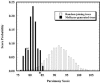
FIG. 2.
TreeClimber-generated probability distribution of MrBayes-generated and neighbor-joining (NJ) phylogenetic trees for the data of McCaig et al. (21). A Scottish soil data set was compared to the distribution of scores obtained from 10,000 random joining trees when there were 137 and 138 samples for each treatment. The asterisk at a parsimony score of 84 indicates the score at which the cumulative random probability was equal to 0.047. When MrBayes-generated trees were used, 98.2% of the trees had a P value less than 0.05 and the P value for the neighbor-joining tree was 0.004.
References
- Begon, M., J. L. Harper, and C. R. Townsend. 1996. Ecology: individuals, populations, and communities, 3rd ed., p. 829-830. Blackwell Science, Malden, MA.
- Chao, A., R. L. Chazdon, R. K. Colwell, and T. J. Shen. 2005. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett. 8:148-159.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical