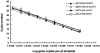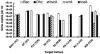Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains - PubMed (original) (raw)
Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains
Kirsti M Ritalahti et al. Appl Environ Microbiol. 2006 Apr.
Abstract
The 16S rRNA gene provides insufficient information to infer the range of chloroorganic electron acceptors used by different Dehalococcoides organisms. To overcome this limitation and provide enhanced diagnostic tools for growth measurements, site assessment, and bioremediation monitoring, a quantitative real-time PCR (qPCR) approach targeting 16S rRNA genes and three Dehalococcoides reductive dehalogenase (RDase) genes with assigned function (i.e., tceA, bvcA, and vcrA) was designed and evaluated. qPCR standard curves generated for the RDase genes by use of genomic DNA from Dehalococcoides pure cultures correlated with standard curves obtained for both Bacteria- and Dehalococcoides-targeted 16S rRNA genes, suggesting that the RDase genes are useful targets for quantitative assessment of Dehalococcoides organisms. RDase gene probe/primer pairs were specific for the Dehalococcoides strains known to carry the diagnostic RDase gene sequences, and the qPCR method allowed the detection of as few as 1 to 20 and quantification of as few as 50 to 100 tceA, bvcA, or vcrA gene targets per PCR volume. The qPCR approach was applied to dechlorinating enrichment cultures, microcosms, and samples from a contaminated site. In characterized enrichment cultures where known Dehalococcoides strains were enumerated, the sum of the three RDase genes equaled the total Dehalococcoides cell numbers. In site samples and chloroethane-dechlorinating microcosms, the sum of the three RDase genes was much less than that predicted by Dehalococcoides-targeted qPCR, totaling 10 to 30% of the total Dehalococcoides cell numbers. Hence, a large number of Dehalococcoides spp. contain as-yet-unidentified RDase genes, indicating that our current understanding of the dechlorinating Dehalococcoides community is incomplete.
Figures
FIG. 1.
qPCR with Dehalococcoides (Dhc*) and Bacteria (Bac*) 16S rRNA gene-targeted primer/probe sets, using Dehalococcoides genomic and plasmid DNA carrying a Dehalococcoides 16S rRNA gene fragment as the template. Dehalococcoides sp. strain BAV1 genomic DNA (diamond) and pBAV1/16S (triangle) interrogated with the _Dehalococcoides_-targeted (open) and universal bacterial (filled) probes were not statistically different under optimized conditions (95% confidence interval, P > 0.05). The dotted line at 2 × 108 gene copies per μl of template indicates the maximum copies per reaction mixture that could be quantified under the reaction conditions tested. The points enclosed in the ellipse (filled circles) indicate values generated with the universal probes and BAV1 plasmid DNA at 56°C, which differed from the standard curves generated at 52°C. The inset shows a comparison of standard curves generated with the bacterial 16S rRNA gene-targeted (Bac*) primer/probe set for Dehalococcoides (open diamond, solid line) and Anaeromyxobacter (filled square, dashed line). The data sets for both organisms were not statistically different (n = 3, P > 0.05).
FIG. 2.
Standard curves for Dehalococcoides 16S rRNA genes and RDase genes. Shown are the _Dehalococcoides_-specific 16S rRNA gene primer/probe set (Dhc*) with pFL2/16S rRNA gene fragment as the template, the _bvcA_-targeted primer/probe set (bvcA*) with pBAV1/bvcA as the template, the _vcrA_-targeted primer/probe set (vcrA*) with pGT/vcrA as the template, and the _tceA_-targeted primer/probe set (tceA*) with pFL2/tceA as the template.
FIG. 3.
Analysis of _Dehalococcoides_-containing cultures by use of the specific qPCR primers and probes. The bar graph depicts three pure Dehalococcoides (Dhc) cultures (BAV1, GT, and FL2), two SuZi Creek (SZ) sediment-derived consortia (enriched with VC or PCE), two 1,2-D-dechlorinating consortia, and the BDI consortium grown on TCE. The provided chlorinated electron acceptor is designated in parentheses. Each bar represents the average of two DNA extractions, each quantified in triplicate (n = 6). Note that the gene copy numbers are given on a log scale; therefore, the heights of the bars are not additive. Bac, Bacteria.
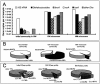
FIG. 4.
qPCR analysis of GW samples collected at a site in Michigan contaminated with chloroethenes and other chlorinated compounds. (A) qPCR data from duplicate microcosms established with site aquifer material and site GW or MM following a 206-day incubation period. Each bar represents the average of duplicate DNA extractions from one microcosm per treatment, each quantified in triplicate (n = 6). Dhc, Dehalococcoides. (B) Pie charts describing the proportions of non-Dehalococcoides and Dehalococcoides 16S rRNA genes in the initial GW and the enrichment cultures. Panel C highlights the shift in known RDase gene distribution prior to and following enrichment in GW or MM.
References
- Abelson, P. H. 1990. Inefficient remediation of ground-water pollution. Science 250:733. - PubMed
- Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. - PubMed
- Aiello, M. R. 2003. Quantitative environmental monitoring of PCE dechlorinators in a contaminated aquifer and PCE-fed bioreactor. M.S. thesis. Michigan State University, East Lansing.
- Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1997. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, Inc., New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous