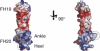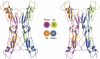Structure of complement factor H carboxyl-terminus reveals molecular basis of atypical haemolytic uremic syndrome - PubMed (original) (raw)
Structure of complement factor H carboxyl-terminus reveals molecular basis of atypical haemolytic uremic syndrome
T Sakari Jokiranta et al. EMBO J. 2006.
Abstract
Factor H (FH) is the key regulator of the alternative pathway of complement. The carboxyl-terminal domains 19-20 of FH interact with the major opsonin C3b, glycosaminoglycans, and endothelial cells. Mutations within this area are associated with atypical haemolytic uremic syndrome (aHUS), a disease characterized by damage to endothelial cells, erythrocytes, and kidney glomeruli. The structure of recombinant FH19-20, solved at 1.8 A by X-ray crystallography, reveals that the short consensus repeat domain 20 contains, unusually, a short alpha-helix, and a patch of basic residues at its base. Most aHUS-associated mutations either destabilize the structure or cluster in a unique region on the surface of FH20. This region is close to, but distinct from, the primary heparin-binding patch of basic residues. By mutating five residues in this region, we show that it is involved, not in heparin, but in C3b binding. Therefore, the majority of the aHUS-associated mutations on the surface of FH19-20 interfere with the interaction between FH and C3b. This obviously leads to impaired control of complement attack on plasma-exposed cell surfaces in aHUS.
Figures
Figure 1
Electrostatic potential of FH19–20 on the surface in two views 90° apart. Positive surface is blue and negative red.
Figure 2
Tetrameric assembly of FH19–20, in stereo view. Each of the monomers is in a different colour. The B and C monomer amino-termini are on top (blue and purple), while the A and D amino-termini are at the bottom (red and green). A schematic view from the top is shown.
Figure 3
Location of aHUS mutations in the FH19–20 structure. (A) A stereo view of FH19–20 with locations of residues mutated in aHUS patients. The mutated residues are coloured according to the putative consequence: brown, disrupting normal folding; green, interference with binding to C3b/C3d; and blue, putative interference with oligomerization. Disulphide bridges are orange. (B) Sequence alignment of the carboxyl-terminal domains of human and murine FH and human FHR-4. The conserved cysteines are shown in black. The residues of FH mutated in aHUS patients are indicated with arrows. Surface-exposed residues of human FH20 and identical or highly homologous residues in murine FH20 or human FHR-4 are shown in gray. Surface-exposed residues shared by these three proteins, and therefore potentially involved in interactions with C3b/C3d, are indicated below the alignment. The site-directed mutants we made in FH19–20 are shown at the bottom.
Figure 4
Heparin affinity chromatography of FH19–20 and mutants. (A) Coomassie blue-stained SDS–PAGE gel of FH19–20 and purified mutants. (B) Western blot of column fractions for wt and the mutants. Each construct was injected onto the heparin column in PBS and eluted using a linear NaCl gradient. The fractions were run on SDS–PAGE and blotted with anti-FH antibody. Fractions collected for Western blotting are indicated with vertical lines; average and minimum/maximum NaCl concentration is shown at the top and the mobility of size markers is indicated on the left.
Figure 5
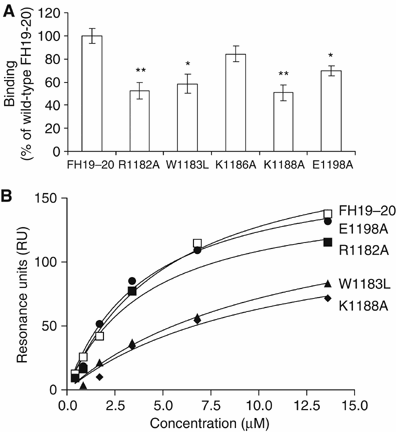
Binding of FH19–20 and the mutant constructs to C3b and C3d. (A) Binding of 125I-labelled C3b to microtiter plate-coupled wt or mutant FH19–20 constructs. The results are shown in comparison to wt FH19–20. Standard deviations are indicated with bars and statistical significance of the difference of each mutant in comparison to wt FH19–20 are indicated by asterisks (*P<0.01 and **P<0.001). (B) SPR analysis (Biacore®) of binding of the fluid-phase FH19–20 constructs to solid-phase C3b. The interaction between the fluid-phase FH19–20 constructs and solid-phase C3b is shown as a function of the fluid-phase construct concentration. Each construct was injected separately to a blank control flow cell and a C3b-coupled flow cell using a flow rate of 30 μl/min and several protein concentrations (3–200 μg/ml).
Figure 6
Docking model of FH19–20 binding to C3d and heparin. (A) The mutations in FH19–20 that have been characterized in aHUS patients are colored as in Figure 3A. On C3d, the FH-interface area is marked with red, while the surface region which interacts with CD21 (Szakonyi et al, 2001) is shown in magenta. (B) The molecules have been dragged apart and rotated away from each other to better visualize the hypothetical interacting regions. (C) Location of a heparin decamer after superimposition of vaccinia complement control protein–heparin complex with FH20. The positively charged residues previously proposed to interact with heparin are in cyan.
Figure 7
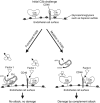
Schematic model of how mutations in FH might affect complement regulation on endothelial cell surfaces. Under normal circumstances FH has sufficient avidity for surface-associated C3b to be able to promote its inactivation into iC3b in concert with membrane-bound complement regulator CD46 (left). Reduced avidity of FH to cell-bound C3b by a mutation in FH20 can result in a failure to efficiently inactivate surface-bound C3b molecules thereby leading to complement-mediated cell damage (right).
Similar articles
- Both domain 19 and domain 20 of factor H are involved in binding to complement C3b and C3d.
Bhattacharjee A, Lehtinen MJ, Kajander T, Goldman A, Jokiranta TS. Bhattacharjee A, et al. Mol Immunol. 2010 May;47(9):1686-91. doi: 10.1016/j.molimm.2010.03.007. Epub 2010 Apr 7. Mol Immunol. 2010. PMID: 20378178 - Mutations of factor H impair regulation of surface-bound C3b by three mechanisms in atypical hemolytic uremic syndrome.
Lehtinen MJ, Rops AL, Isenman DE, van der Vlag J, Jokiranta TS. Lehtinen MJ, et al. J Biol Chem. 2009 Jun 5;284(23):15650-8. doi: 10.1074/jbc.M900814200. Epub 2009 Apr 7. J Biol Chem. 2009. PMID: 19351878 Free PMC article. - Disturbed sialic acid recognition on endothelial cells and platelets in complement attack causes atypical hemolytic uremic syndrome.
Hyvärinen S, Meri S, Jokiranta TS. Hyvärinen S, et al. Blood. 2016 Jun 2;127(22):2701-10. doi: 10.1182/blood-2015-11-680009. Epub 2016 Mar 22. Blood. 2016. PMID: 27006390 - Complement activation in diseases presenting with thrombotic microangiopathy.
Meri S. Meri S. Eur J Intern Med. 2013 Sep;24(6):496-502. doi: 10.1016/j.ejim.2013.05.009. Epub 2013 Jun 4. Eur J Intern Med. 2013. PMID: 23743117 Review. - Factor H family proteins: on complement, microbes and human diseases.
Zipfel PF, Skerka C, Hellwage J, Jokiranta ST, Meri S, Brade V, Kraiczy P, Noris M, Remuzzi G. Zipfel PF, et al. Biochem Soc Trans. 2002 Nov;30(Pt 6):971-8. doi: 10.1042/bst0300971. Biochem Soc Trans. 2002. PMID: 12440956 Review.
Cited by
- The major autoantibody epitope on factor H in atypical hemolytic uremic syndrome is structurally different from its homologous site in factor H-related protein 1, supporting a novel model for induction of autoimmunity in this disease.
Bhattacharjee A, Reuter S, Trojnár E, Kolodziejczyk R, Seeberger H, Hyvärinen S, Uzonyi B, Szilágyi Á, Prohászka Z, Goldman A, Józsi M, Jokiranta TS. Bhattacharjee A, et al. J Biol Chem. 2015 Apr 10;290(15):9500-10. doi: 10.1074/jbc.M114.630871. Epub 2015 Feb 6. J Biol Chem. 2015. PMID: 25659429 Free PMC article. - Zinc binding to the Tyr402 and His402 allotypes of complement factor H: possible implications for age-related macular degeneration.
Nan R, Farabella I, Schumacher FF, Miller A, Gor J, Martin AC, Jones DT, Lengyel I, Perkins SJ. Nan R, et al. J Mol Biol. 2011 May 13;408(4):714-35. doi: 10.1016/j.jmb.2011.03.006. Epub 2011 Mar 17. J Mol Biol. 2011. PMID: 21396937 Free PMC article. - Secondary focal and segmental glomerulosclerosis associated with single-nucleotide polymorphisms in the genes encoding complement factor H and C3.
Sethi S, Fervenza FC, Zhang Y, Smith RJ. Sethi S, et al. Am J Kidney Dis. 2012 Aug;60(2):316-21. doi: 10.1053/j.ajkd.2012.04.011. Epub 2012 May 16. Am J Kidney Dis. 2012. PMID: 22594991 Free PMC article. - Structural analysis of the C-terminal region (modules 18-20) of complement regulator factor H (FH).
Morgan HP, Mertens HD, Guariento M, Schmidt CQ, Soares DC, Svergun DI, Herbert AP, Barlow PN, Hannan JP. Morgan HP, et al. PLoS One. 2012;7(2):e32187. doi: 10.1371/journal.pone.0032187. Epub 2012 Feb 28. PLoS One. 2012. PMID: 22389686 Free PMC article. - Microbes bind complement inhibitor factor H via a common site.
Meri T, Amdahl H, Lehtinen MJ, Hyvärinen S, McDowell JV, Bhattacharjee A, Meri S, Marconi R, Goldman A, Jokiranta TS. Meri T, et al. PLoS Pathog. 2013;9(4):e1003308. doi: 10.1371/journal.ppat.1003308. Epub 2013 Apr 18. PLoS Pathog. 2013. PMID: 23637600 Free PMC article.
References
- Alitalo A, Meri T, Chen T, Lankinen H, Cheng ZZ, Jokiranta TS, Seppala IJ, Lahdenne P, Hefty PS, Akins DR, Meri S (2004) Lysine-dependent multipoint binding of the Borrelia burgdorferi virulence factor outer surface protein E to the C terminus of factor H. J Immunol 172: 6195–6201 - PubMed
- Aslam M, Perkins SJ (2001) Folded-back solution structure of monomeric factor H of human complement by synchrotron X-ray and neutron scattering, analytical ultracentrifugation and constrained molecular modelling. J Mol Biol 309: 1117–1138 - PubMed
- Barlow PN, Steinkasserer A, Norman DG, Kieffer B, Wiles AP, Sim RB, Campbell ID (1993) Solution structure of a pair of complement modules by nuclear magnetic resonance. J Mol Biol 232: 268–284 - PubMed
- Blackmore TK, Hellwage J, Sadlon TA, Higgs N, Zipfel PF, Ward HM, Gordon DL (1998) Identification of the second heparin-binding domain in human complement factor H. J Immunol 160: 3342–3348 - PubMed
- Caprioli J, Bettinaglio P, Zipfel PF, Amadei B, Daina E, Gamba S, Skerka C, Marziliano N, Remuzzi G, Noris M (2001) The molecular basis of familial hemolytic uremic syndrome: mutation analysis of factor H gene reveals a hot spot in short consensus repeat 20. J Am Soc Nephrol 12: 297–307 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous