ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks - PubMed (original) (raw)
ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks
Kristina Trenz et al. EMBO J. 2006.
Abstract
Ataxia-telangiectasia mutated (ATM), ataxia-telangiectasia Rad3-related (ATR) and the Mre11/Rad50/Nbs1 complex ensure genome stability in response to DNA damage. However, their essential role in DNA metabolism remains unknown. Here we show that ATM and ATR prevent accumulation of DNA double-strand breaks (DSBs) during chromosomal replication. Replicating chromosomes accumulate DSBs in Xenopus laevis egg extracts depleted of ATM and ATR. Addition of ATM and ATR proteins to depleted extracts prevents DSB accumulation by promoting restart of collapsed replication forks that arise during DNA replication. We show that collapsed forks maintain MCM complex but lose Pol epsilon, and that Pol epsilon reloading requires ATM and ATR. Replication fork restart is abolished in Mre11 depleted extracts and is restored by supplementation with recombinant human Mre11/Rad50/Nbs1 complex. Using a novel fluorescence resonance energy transfer-based technique, we demonstrate that ATM and ATR induce Mre11/Rad50/Nbs1 complex redistribution to restarting forks. This study provides direct biochemical evidence that ATM and ATR prevent accumulation of chromosomal abnormalities by promoting Mre11/Rad50/Nbs1 dependent recovery of collapsed replication forks.
Figures
Figure 1
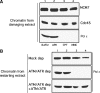
ATM and ATR prevent DSB accumulation during chromosomal DNA replication. (A) DSBs are detected by the TUNEL assay measuring incorporation of α-32P-labeled dGTP into genomic DNA in the presence of TdT (see Supplementary Materials and methods). Upper panel: Extracts supplemented with Buffer (+Buffer). Lower panel: Extracts supplemented with recombinant Flag-ATM and Flag-ATR (+ATM/ATR). Labeling of postreplicative chromatin isolated from untreated extracts (Control, lanes 1 and 7), mock-depleted extracts (Mock dep, lanes 2 and 8), an extract supplemented with 5 mM caffeine (Caff) and 4 ng/μl of Geminin (GEM) (lane 3), ATM and ATR depleted extracts (ATM and ATR dep) treated with 4 ng/μl of Geminin (lanes 4 and 9), an extract treated with 5 mM caffeine (Caff, lane 5) and ATM and ATR depleted extracts (ATM/ATR dep, lanes 6 and 10). (B) TUNEL assay on intact nuclei. Postreplicative nuclei stained with DAPI (upper panels) or with Fluorescein-dUTP in the presence of TdT (lower panels) (see Supplementary Materials and methods). Nuclei were isolated from an untreated extract (Control), an extract treated with 5 mM caffeine (Caff), an extract treated with 5 mM caffeine and 500 μM Roscovitine (Caff+High Rosc) or an extract treated with 5 mM caffeine and 4 ng/μl of Geminin (Caff+GEM). (C) Quantification of the TUNEL assay. Labeling of postreplicative chromatin isolated from a control extract (Control), a mock-depleted extracts (Mock dep), an extract supplemented with 5 mM caffeine and 4 ng/μl of Geminin (Caff+GEM), an extract supplemented with 5 mM caffeine and 500 μM Roscovitine (Caff+High Rosc), an extract supplemented with 5 mM caffeine (Caff), an ATM and ATR depleted extract treated with 4 ng/μl of Geminin (ATM/ATR dep+GEM), an ATM and ATR depleted extract supplemented with 500 μM Roscovitine (ATM/ATR dep+High Rosc), an ATM and ATR depleted extract (ATM/ATR dep), an ATR depleted extract (ATR dep), an ATM depleted extract (ATM dep), an ATM and ATR depleted extract supplemented with 5 μM Roscovitine (ATM/ATR dep+Low Rosc), an ATM and ATR depleted extract supplemented with Flag-ATM and Flag-ATR (ATM/ATR dep+ATM/ATR) and an ATM and ATR depleted extract supplemented with catalytically inactive Flag-ATM-KD and Flag-ATR-KD proteins (ATM/ATR dep+ATM/ATR KD). (D) Immune and peptide mediated depletion of ATM and ATR. Western blot analysis with anti-Xenopus ATM (upper panel) and anti-Xenopus ATR (lower panel) of control extracts (Control), mock depleted extracts (Mock dep), an extract depleted with biotinylated C-terminal peptide of Xenopus Nbs1 (ATM dep) and an extract depleted with anti-ATR antibodies (ATR dep). (E) Reconstitution of ATM and ATR activity in extracts. The activity of ATM and ATR protein kinases was monitored by incorporation of 32P from γ-32P-labeled ATP into GST fused to the C-terminal peptide of histone H2AX (GST-H2AX). 32P incorporation into GST- H2AX in the absence or in the presence of linear DNA at 50 ng/μl (+DSBs) was monitored in an extract incubated with buffer (Buffer), with 5 mM caffeine (Caff), in a mock depleted extract (Mock dep), in an ATM and ATR depleted extract (ATM/ATR dep), in an ATM and ATR depleted extract supplemented with catalytically inactive Flag-ATM and Flag-ATR proteins (ATM/ATR dep+ATM/ATR KD) and in an ATM and ATR depleted extract supplemented with recombinant Flag-ATM and Flag-ATR (ATM/ATR dep+ATM/ATR).
Figure 2
ATM and ATR inhibit DSB accumulation in the presence of damaged replication forks. DSBs detected by TUNEL labeling of postreplicative chromatin isolated after 120-min incubation in control extracts (A, Control), in extracts supplemented at 0 min with 4 ng/μl of Geminin (B, GEM), 500 μM Roscovitine (C, High Rosc) or 5 μM Roscovitine (D, Low Rosc). Extracts were supplemented at 60 min from nuclei addition with buffer (Buffer), with 55 μM CPT or with 300 μM MMC. Extracts were mock depleted (gray bars), supplemented with 5 mM caffeine (black bars), ATM and ATR depleted (white bars) or untreated (striped bars).
Figure 3
ATM and ATR promote restart of collapsed replication forks. (A) Experimental approach to dissect the role of ATM and ATR in the restart of collapsed replication forks. Chromatin replication is initiated in a first interphase extract (damaging extract). Replication fork progression is then blocked by addition of APH, CPT or MMC 60 min after nuclei addition to extracts in the presence of 5 mM caffeine and 500 μM Roscovitine. At 90 min after nuclei addition chromatin with damaged forks is isolated and incubated in a second interphase extract (restarting extract) made incompetent for origin firing and origin assembly by addition of 500 μM Roscovitine and 4 ng/μl Geminin. Different restarting extracts were used as stated in the figure. (B) DNA replication in damaging extracts supplemented at 60 min after nuclei addition with 5 mM caffeine, 500 μM Roscovitine and buffer (Buffer, lane 1), 40 μM APH (lane 2), 55 μM CPT (lane 3) or 300 μM MMC (lane 4). Replication was pulse labeled with α-32P-labeled dGTP from 60 min to 90 min from nuclei addition. (C) DNA replication in restarting extracts. Chromatin preincubated in damaging extracts was isolated after 90 min and then transferred to restarting extracts that contained 500 μM Roscovitine and 4 ng/μl Geminin. Replication was monitored with α-32P-labeled dGTP for 90 min. DNA was then isolated and run on neutral agarose gel. The different panels show replication of chromatin isolated from damaging extracts treated with Buffer (lanes 5 and 9), 40 μM APH (lanes 6 and 10), 55 μM CPT (lanes 7, 11, 13, 15, 17, 19, 21) or 300 μM MMC (lanes 8, 12, 14, 16, 18, 20, 22) and transferred to restarting extracts. Restarting extracts were mock depleted (Mock dep), ATM and ATR depleted (ATM/ATR dep), treated with 5 mM caffeine (Caff), ATM and ATR depleted and supplemented with recombinant Flag-ATM and Flag-ATR (ATM/ATR dep+ATM/ATR), ATM and ATR depleted and supplemented with catalytically inactive Flag-ATM and Flag-ATR proteins (ATM/ATR dep+ATM/ATR KD), ATR depleted (ATR dep) or ATM depleted (ATM dep). Note that the amount of DNA loaded on the gel in panel C is two-fold more than the amount of DNA loaded on the gel in panel B (see Supplementary Materials and methods). (D) Quantification of DNA replication in restarting extracts. Extracts were supplemented with buffer (white bars), 40 μM APH (black bars), 55 μM CPT (gray bars) or 300 μM MMC (striped bars) treated chromatin. Graph shown represents a typical experiment. (E) Nascent single strand DNA synthesis in restarting extracts. Chromatin was preincubated in damaging extracts treated with buffer (Buffer, lane 1), 40 μM APH (lane 2), 55 μM CPT (lane 3) or 300 μM MMC (lane 4), isolated and then transferred to restarting extracts. Replication was monitored for 60 min in the presence of α-32P-labeled dGTP. DNA was isolated and run on an alkaline agarose gel.
Figure 4
ATM and ATR promote reloading of Pol ɛ onto replication forks. (A) Western blot analysis of chromatin isolated from damaging extracts supplemented at 60 min from nuclei addition with 5 mM Caffeine, 500 μM Roscovitine+buffer (Buffer, lane 1), 40 μM APH (lane 2), 55 μM CPT (lane 3) or 300 μM MMC (lane 4) using anti-MCM7 antibodies (MCM7), anti-Cdc45 antibodies (Cdc45) or anti-Pol ɛ antibodies (Pol ɛ). (B) Western blot analysis using anti-Pol ɛ antibodies of chromatin isolated from damaging extracts that had been incubated at 60 min after sperm nuclei addition with 5 mM caffeine and 500 μM Roscovitine+buffer (Buffer, lane 1), 40 μM APH (lane 2), 55 μM CPT (lane 3) or 300 μM MMC (lane 4) and transferred to a mock depleted extract (Mock dep), an ATM and ATR depleted restarting extract (ATM/ATR dep) or an ATM and ATR depleted extract supplemented with recombinant Flag-ATM and Flag-ATR (ATM/ATR dep+ATM/ATR).
Figure 5
Mre11 complex is required for ATM and ATR-dependent fork restart. (A) DNA replication in restarting extracts. Chromatin was isolated from Mre11 depleted damaging extracts treated with 5 mM caffeine and 500 μM Roscovitine+40 μM APH (lane 1), 55 μM CPT (lanes 2, 4 and 6) or 300 μM MMC (lanes 3, 5 and 7) and then transferred to restarting extracts that were Mre11 depleted (Mre11 dep), Mre11 depleted supplemented with 500 nM of recombinant Mre11/Rad50/Nbs1 complex (Mre11 dep+Mre11/Rad50/Nbs1) or Mre11 depleted supplemented with Flag-ATR and active Flag-ATM proteins (Mre11 dep+ATM* and ATR). Asterisk (*) indicates that ATM has been isolated from irradiated cells (see Supplementary Materials and methods). Replication was monitored for 90 min in the presence of α-32P-labeled dGTP. DNA was then isolated and run on an agarose gel. (B) Quantification of DNA replication in Mre11 depleted restarting extracts. Extracts were supplemented with 40 μM APH (black bars), 55 μM CPT (gray bars) or 300 μM MMC (striped bars) treated chromatin. Graph shown represents a typical experiment.
Figure 6
ATM and ATR promote redistribution of Mre11 complex to restarting replication forks. (A) Chromatin replication is initiated in damaging extracts. At 55 min after addition of sperm nuclei extracts are supplemented with Cy5-dCTP. At 60 min after nuclei addition extracts are supplemented with 40 μM APH, 55 μM CPT or 300 μM MMC. Cy5-dCTP labeled chromatin is then isolated and incubated in restarting extracts that were mock depleted (Mock dep), treated with 5 mM caffeine (Caff), ATM and ATR depleted (ATM/ATR dep) or ATM and ATR depleted supplemented with recombinant Flag-ATM and Flag-ATR (ATM/ATR dep+ATM/ATR). Eu3+-Mre11 labeled complex is added to restarting extract at 0 min or at 45 min from chromatin addition. Chromatin is isolated after 15 min and subjected to fluorometry. (B) Chromatin isolated from restarting extract is excited at 340 nm. Emission is monitored at 618 and 665 nm. Emission at 665 nm is the result of FRET between Eu3+-Mre11 localized in the range of ∼10 nm from Cy5-dCTP labeled DNA. (C) 665/618 nm emission ratio of chromatin preincubated in Mre11 depleted damaging extracts treated with CPT (gray bars), MMC (black bars) or APH (white bars). Cy5-dCTP labeled chromatin was isolated and transferred to restarting extracts supplemented with Europium labeled Mre11 complex at 0 (Eu3+-Mre11 at 0′) or at 45 min (Eu3+-Mre11 at 45′) from chromatin addition. As control nonlabeled Mre11 complex was added at 0 min from chromatin addition (Mre11 at 0′). Chromatin was then re-isolated from restarting extracts 15 min after addition of Eu3+-Mre11 and 665/618 nm emission ratio was measured in a fluorometer. Restarting extracts were mock depleted control, supplemented with caffeine (Caff), ATM and ATR depleted (ATM/ATR dep) or ATM and ATR depleted and supplemented with Flag-ATM and Flag-ATR (ATM/ATR dep+ATM/ATR). As a control, CPT, MMC or APH treated chromatin that did not contain Cy5-dCTP was transferred to restarting extracts supplemented with Eu3+-Mre11 complex at 0 min from chromatin addition (No Cy5-dCTP). (D) Western blot analysis of chromatin bound Mre11. Chromatin was preincubated in Mre11 depleted damaging extract containing 55 μM CPT and transferred to restarting extracts that were mock depleted buffer, ATM and ATR depleted (ATM/ATR dep), ATM and ATR depleted+Flag-ATM and Flag-ATR (ATM/ATR dep+ATM/ATR) or supplemented with 5 mM caffeine (Caff). For Western blot anti-Xenopus Mre11 antibodies were used.
Figure 7
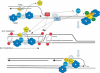
Multiple roles for ATM, ATR and Mre11 complex in promoting genome stability during DNA replication: ATM and ATR prevent DSB accumulation during normal or challenged DNA replication by regulating origin firing; by coordinating redistribution of Mre11 complex to collapsed replication forks; by stimulating restart of the collapsed replication fork and by promoting reloading of Pol ɛ onto recovering forks.
References
- Caldecott KW (2003) XRCC1 and DNA strand break repair. DNA Repair (Amst) 2: 955–969 - PubMed
- Caldecott KW (2004) DNA single-strand breaks and neurodegeneration. DNA Repair (Amst) 3: 875–882 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous