Juxtamembranous aspartic acid in Insig-1 and Insig-2 is required for cholesterol homeostasis - PubMed (original) (raw)
Juxtamembranous aspartic acid in Insig-1 and Insig-2 is required for cholesterol homeostasis
Yi Gong et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Insig-1 and Insig-2 are closely related proteins of the endoplasmic reticulum (ER) that mediate feedback control of cholesterol synthesis by sterol-dependent binding to the following two membrane proteins: the escort protein Scap, thus preventing proteolytic processing of sterol regulatory element-binding proteins; and the cholesterol biosynthetic enzyme 3-hydroxy-3-methylglutaryl CoA reductase, thus inducing the ubiquitination and ER-associated degradation of the enzyme. Here, we report that the conserved Asp-205 in Insig-1, which abuts the fourth transmembrane helix at the cytosolic side of the ER membrane, is essential for its dual function. When Asp-205 was mutated to alanine, the mutant Insig-1 lost the ability to bind to Scap and, thus, was unable to suppress the cleavage of sterol regulatory element-binding proteins. The mutant Insig-1 was ineffective also in accelerating sterol-stimulated degradation of 3-hydroxy-3-methylglutaryl CoA reductase. Alanine substitution of the corresponding aspartic acid in Insig-2 produced the same dual defects. These studies identify a single amino acid residue that is crucial for the function of Insig proteins in regulating cholesterol homeostasis in mammalian cells.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Fig. 1.
Sequence alignment of Insig proteins and membrane topology of human Insig-1. (A) Sequence alignment of the membrane domain of Insig proteins was performed by
clustalw
program (DNASTAR, Madison, WI). Residues identical to human Insig-1 are shown in yellow, and two conserved motifs are indicated by the bars. GenBank accession nos. for human Insig-1, human Insig-2, hamster Insig-1, Xenopus Insig-1, zebrafish Insig-1, and Schizosaccharomyces pombe Insig are AY112745, AF527632, AF527628, BC076862, AF527626, and CAB41653, respectively. (B) Proposed membrane topology of Insig-1 (13). Asp-205 is shown in black. Residues other than Asp-205 in the two conserved motifs are shown in orange. The residue in human Insig-2 corresponding to Asp-205 in human Insig-1 is Asp-149 (9).
Fig. 2.
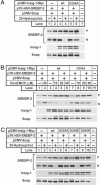
Mutant Insig-1(D205A) is defective in suppressing SREBP-2 cleavage. On day 0, SRD-13A cells were set up at 3.5 × 105 cells per 60-mm dish. On day 2, cells in each dish were transfected with 0.4 μg of pCMV-Scap, 2 μg of pTK-HSV-SREBP-2, and 0.1 μg of WT or 0.3 μg of mutant pCMV-Insig1-Myc, as indicated. On day 3, cells were switched to medium C supplemented with 1% hydroxypropyl-β-cyclodextrin and incubated for 1 h at 37°C. Cells were then washed twice with PBS and switched to medium C supplemented either with (+) or without (−) 1 μg/ml 25-HC (A and C) or the indicated concentration of cholesterol complexed to methyl-β-cyclodextrin (B). After incubation for 5 h, cells were harvested, fractionated into membrane and nuclear extracts, and subjected to SDS/PAGE and immunoblot analysis. Membrane fractions were immunoblotted with anti-HSV, IgG-9E10, and IgG-R139 to detect precursor form of SREBP-2 (P), Insig-1, and Scap, respectively. The nuclear extracts were immunoblotted with anti-HSV to detect the nuclear form of SREBP-2. Filters were exposed to film for 5–30 s. Chol/MCD, cholesterol complexed to methyl-β-cyclodextrin.
Fig. 3.
Mutant Insig-1(D205A) does not bind to Scap but does bind to VAP proteins. (A) On day 0, SRD-13A cells were set up at 3.5 × 105 cells per 60-mm dish. On day 2, cells in each dish were transfected with 0.5 μg of pCMV-Scap and 0.1 μg of WT or 0.25 μg of D205A mutant pCMV-Insig1-Myc, as indicated. At 6 h after the transfection, cells were changed to sterol-depleting medium C. On day 3, cells were switched to fresh medium C in the absence (−) or presence (+) of 0.3 μg/ml 25-HC. After incubation for 5 h, cells were harvested, lysed, and immunoprecipitated (IP) with polyclonal anti-Myc to precipitate Insig-1. Pellets (representing 0.25 dish of cells) and supernatants (representing 0.05 dish of cells) of the immunoprecipitation were subjected to SDS/PAGE and immunoblot (IB) analysis. Filters were exposed to film for 2–60 s. (B) On day 0, CHO-7 cells were set up at 7 × 105 cells per 60-mm dish. On day 1, cells in each dish were transfected with 1 μg of pCMV-VAP-A-HA, 0.3 μg of pCMV-VAP-B-HA, 0.1 μg of WT, or 0.2 μg of D205A mutant pCMV-Insig1-Myc, as indicated. On day 3, cells were harvested, lysed, and immunoprecipitated (IP) with polyclonal anti-HA to precipitate VAP proteins. Pellets (representing 0.25 dish of cells) and supernatants (representing 0.05 dish of cells) were subjected to SDS/PAGE and immunoblot analysis. Filters were exposed to film for 1–60 s.
Fig. 4.
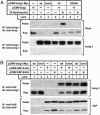
Mutant Insig-1(D205A) is not stabilized by Scap. On day 0, SRD-13A cells were set up at 3.5 × 105 cells per 60-mm dish. On day 2, cells were transfected with 1 μg of pTK-Scap and 0.2 μg of WT or D205A mutant pTK-Insig1-Myc, as indicated. At 6 h after transfection, cells were changed to sterol-depleting medium C. On day 3, cells were switched to fresh medium C in the absence (−) or presence (+) of 0.1 μg/ml 25-HC, with or without the addition of 10 μM MG-132. After incubation for 5 h, cells were harvested, and cell lysates were subjected to SDS/PAGE and immunoblot analysis with IgG-9E10 and IgG-R139 to detect Insig-1 and Scap, respectively. Filters were exposed to film for 10–60 s.
Fig. 5.
Mutant Insig-1(D205A) is defective in accelerating the degradation of HMG CoA reductase in the presence of 25-HC or lanosterol. On day 0, CHO-K1 cells were set up at 5 × 105 cells per 60-mm dish. On day 1, cells were transfected in A with 1 μg of pCMV-HMG-Red-T7(TM1–8) and the indicated amount of WT or D205A mutant pCMV-Insig1-Myc and in B with 1 μg of pCMV-HMG-Red-T7(TM1–8) and either 0.1 μg of WT or 0.3 μg of D205A mutant pCMV-Insig1-Myc. At 6 h after transfection, cells were changed to sterol-depleting medium C. On day 3, cells were switched to fresh medium C in the absence (−) or presence (+) of either 1 μg/ml 25-HC plus 10 mM mevalonate (A) or the indicated concentration of lanosterol plus 10 mM mevalonate (B). After incubation for 5 h, cells were harvested, and membrane fractions were subjected to SDS/PAGE and immunoblot analysis with anti-T7 mAb and IgG-9E10 to detect HMG CoA reductase and Insig-1, respectively. Filters were exposed to film for 5–30 s.
Fig. 6.
Mutant Insig-2(D149A) is defective in suppressing SREBP-2 cleavage and accelerating the degradation of HMG CoA reductase. (A) SRD-13A cells were set up, treated, and analyzed in the same manner as described in Fig. 2_A_ except that 1 μg of WT or 3 μg of mutant pCMV-Insig2-Myc was transfected. (B) CHO-K1 cells were set up, treated, and analyzed in the same manner as described in Fig. 5. Filters were exposed to film for 5–30 s.
Similar articles
- Dual functions of Insig proteins in cholesterol homeostasis.
Dong XY, Tang SQ, Chen JD. Dong XY, et al. Lipids Health Dis. 2012 Dec 18;11:173. doi: 10.1186/1476-511X-11-173. Lipids Health Dis. 2012. PMID: 23249523 Free PMC article. Review. - Isolation of sterol-resistant Chinese hamster ovary cells with genetic deficiencies in both Insig-1 and Insig-2.
Lee PC, Sever N, Debose-Boyd RA. Lee PC, et al. J Biol Chem. 2005 Jul 1;280(26):25242-9. doi: 10.1074/jbc.M502989200. Epub 2005 May 2. J Biol Chem. 2005. PMID: 15866869 - Intramembrane glycine mediates multimerization of Insig-2, a requirement for sterol regulation in Chinese hamster ovary cells.
Lee PC, DeBose-Boyd RA. Lee PC, et al. J Lipid Res. 2010 Jan;51(1):192-201. doi: 10.1194/jlr.M900336-JLR200. J Lipid Res. 2010. PMID: 19617589 Free PMC article. - Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER.
Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS. Yang T, et al. Cell. 2002 Aug 23;110(4):489-500. doi: 10.1016/s0092-8674(02)00872-3. Cell. 2002. PMID: 12202038 - Insulin-induced gene: a new regulator in lipid metabolism.
Dong XY, Tang SQ. Dong XY, et al. Peptides. 2010 Nov;31(11):2145-50. doi: 10.1016/j.peptides.2010.07.020. Epub 2010 Sep 15. Peptides. 2010. PMID: 20817058 Review.
Cited by
- A tagging SNP in INSIG2 is associated with obesity-related phenotypes among Samoans.
Deka R, Xu L, Pal P, Toelupe PT, Laumoli TS, Xi H, Zhang G, Weeks DE, McGarvey ST. Deka R, et al. BMC Med Genet. 2009 Dec 22;10:143. doi: 10.1186/1471-2350-10-143. BMC Med Genet. 2009. PMID: 20028541 Free PMC article. - INSIG2 gene polymorphism is associated with increased subcutaneous fat in women and poor response to resistance training in men.
Orkunoglu-Suer FE, Gordish-Dressman H, Clarkson PM, Thompson PD, Angelopoulos TJ, Gordon PM, Moyna NM, Pescatello LS, Visich PS, Zoeller RF, Harmon B, Seip RL, Hoffman EP, Devaney JM. Orkunoglu-Suer FE, et al. BMC Med Genet. 2008 Dec 23;9:117. doi: 10.1186/1471-2350-9-117. BMC Med Genet. 2008. PMID: 19105843 Free PMC article. - Insig2 is associated with colon tumorigenesis and inhibits Bax-mediated apoptosis.
Li CG, Gruidl M, Eschrich S, McCarthy S, Wang HG, Alexandrow MG, Yeatman TJ. Li CG, et al. Int J Cancer. 2008 Jul 15;123(2):273-282. doi: 10.1002/ijc.23510. Int J Cancer. 2008. PMID: 18464289 Free PMC article. - Glucagon regulates hepatic lipid metabolism via cAMP and Insig-2 signaling: implication for the pathogenesis of hypertriglyceridemia and hepatic steatosis.
Wang H, Zhao M, Sud N, Christian P, Shen J, Song Y, Pashaj A, Zhang K, Carr T, Su Q. Wang H, et al. Sci Rep. 2016 Sep 1;6:32246. doi: 10.1038/srep32246. Sci Rep. 2016. PMID: 27582413 Free PMC article. - Dual functions of Insig proteins in cholesterol homeostasis.
Dong XY, Tang SQ, Chen JD. Dong XY, et al. Lipids Health Dis. 2012 Dec 18;11:173. doi: 10.1186/1476-511X-11-173. Lipids Health Dis. 2012. PMID: 23249523 Free PMC article. Review.
References
- Goldstein J. L., DeBose-Boyd R. A., Brown M. S. Cell. 2006;124:35–46. - PubMed
- Goldstein J. L., Brown M. S. Nature. 1990;343:425–430. - PubMed
- Song B.-L., Sever N., DeBose-Boyd R. A. Mol. Cell. 2005;19:829–840. - PubMed
- Ravid T., Doolman R., Avner R., Harats D., Roitelman J. J. Biol. Chem. 2000;275:35840–35847. - PubMed
- Sever N., Yang T., Brown M. S., Goldstein J. L., DeBose-Boyd R. A. Mol. Cell. 2003;11:25–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases