Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: a coordinated mechanism for severing and capping filaments - PubMed (original) (raw)
Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: a coordinated mechanism for severing and capping filaments
Kyoko Okada et al. Mol Biol Cell. 2006 Jul.
Abstract
Rapid turnover of actin structures is required for dynamic remodeling of the cytoskeleton and cell morphogenesis, but the mechanisms driving actin disassembly are poorly defined. Cofilin plays a central role in promoting actin turnover by severing/depolymerizing filaments. Here, we analyze the in vivo function of a ubiquitous actin-interacting protein, Aip1, suggested to work with cofilin. We provide the first demonstration that Aip1 promotes actin turnover in living cells. Further, we reveal an unanticipated role for Aip1 and cofilin in promoting rapid turnover of yeast actin cables, dynamic structures that are decorated and stabilized by tropomyosin. Through systematic mutagenesis of Aip1 surfaces, we identify two well-separated F-actin-binding sites, one of which contributes to actin filament binding and disassembly specifically in the presence of cofilin. We also observe a close correlation between mutations disrupting capping of severed filaments in vitro and reducing rates of actin turnover in vivo. We propose a model for balanced regulation of actin cable turnover, in which Aip1 and cofilin function together to "prune" tropomyosin-decorated cables along their lengths. Consistent with this model, deletion of AIP1 rescues the temperature-sensitive growth and loss of actin cable defects of tpm1Delta mutants.
Figures
Figure 1.
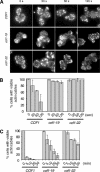
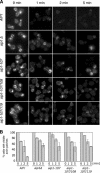
Reduced rates of actin cable turnover in cof1 mutant cells. Wild-type (COF1) and mutant (cof1-19, cof1-22) yeast cells were incubated with 20 μM Latrunculin A (Lat-A). Samples of cells were removed at the indicated time points, fixed, stained with anti-actin antibodies, and scored for visible actin cables. (A) Representative cell staining. (B and C) Percent cells with visible cables remaining after Lat-A treatment for shorter (B) or longer (C) incubation periods. More than 200 cells were counted for each time point, and the columns are an average of two independent experiments. Error bars, SD.
Figure 2.
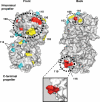
Surfaces on Aip1 required for its in vivo function. Mutated residues were modeled on the crystal structure of S. cerevisiae Aip1 (1PI6) with mutant allele numbers indicated (Table 1). The majority of conserved surface residues map to the “front” face of Aip1 (see Supplementary Figure S1 for sequence alignment). The color scheme for the mutated residues is based on the severity of mutant phenotypes (Table 1): Cyan, pseudowild type; yellow, moderately impaired; red, severely impaired. The two F-actin-binding sites identified (Figure 4) are circled by thick dotted lines. One additional surface that is important for Aip1 in vivo function (disrupted by aip1-109) is circled by a thin dotted line. The inset shows an enlarged view from a different angle of the site mutated in aip1-119. Note that the residues mutated in aip1-119 (rendered in red) protrude from the surface of the β-propeller. Molecular graphics were generated using PyMOL software (DeLano Scientific, South San Francisco, CA).
Figure 3.
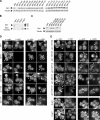
Actin cytoskeleton organization and Aip1 and cofilin localization in wild-type and mutant aip1 cells. (A–C) Aip1 protein levels in wild-type and mutant aip1 cells. Whole cell extracts from integrated wild-type and mutant strains were immunoblotted with anti-Aip1 and anti-tubulin antibodies. (D and E) Cells with the designated genotypes were double-labeled with anti-actin and anti-Aip1 antibodies (left panels) or anti-actin and anti-cofilin antibodies (right panels). Arrowheads point to actin cables; arrows point to actin patches.
Figure 4.
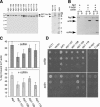
Actin- and cofilin-interaction of purified wild-type and mutant Aip1 proteins. (A) Coomassie-stained gel of purified proteins. Lower arrow points to Aip1 lacking GST tag; upper arrow points to residual uncleaved GST-Aip1. Note that Aip1-119 has retarded migration compared with wild-type Aip1. (B) Cosedimentation of 0.5 μM wild-type Aip1 with 2 μM rabbit skeletal muscle F-actin (RMA) in the presence and absence of 3 μM cofilin; supernatants (S) and pellets (P) were analyzed on Coomassie-stained gels. (C) Comparison of wild-type and mutant Aip1 cosedimentation with RMA, with conditions same as above, in the presence (bottom panel) and absence (top panel) of cofilin (n = 3). Error bars, SD. (D) Two-hybrid interactions of wild-type and mutant Aip1 with cofilin (top) and actin (bottom). Cultures of diploid strains carrying Aip1-DBD bait plasmids and cofilin-AD or actin-AD prey plasmids were serially diluted and plated on triple selective media and 10 mM 3-AT.
Figure 5.
Activities of purified wild-type and mutant Aip1 proteins. (A) Concentration-dependent effects of wild-type Aip1 on actin filament net disassembly. Preassembled yeast F-actin, 4 μM, was incubated at 25°C for 20 min in the presence or absence of 6 μM cofilin and the indicated concentrations of Aip1. The reactions were centrifuged, and the pellets and supernatants were analyzed on Coomassie-stained gels. (B) Concentration-dependent effects of wild-type and mutant Aip1 proteins on actin filament net disassembly. Reactions were carried out as in A. Results from three independent experiments were quantified by densitometry and graphed: wild-type Aip1 (■), Aip1-107 (◇), Aip1-108 (▵), Aip1-109 (○) Aip1-119 (□). Error bars, SD. (C) Cofilin-dependent capping and net disassembly of actin filaments by Aip1. Preassembled yeast F-actin, 4 μM, was incubated for 4 h at 25°C with and without 100 nM Aip1 and/or 400 nM cofilin and/or 6 μM profilin and then centrifuged, and the pellets and supernatants were analyzed on Coomassie-stained gels. (D) Effects of wild-type and mutant Aip1 proteins on cofilin-dependent capping and net disassembly of actin filaments with reactions performed as in C. F-actin levels in the pellet were quantified by densitometry and graphed as a percentage of F-actin in the pellet for wild-type Aip1. n = 2 for Aip1, Aip1-103, Aip1-104, Aip1-106, Aip1-111, Aip1-113, Aip1-114, Aip1-115, and Aip1-118. n = 3 for Aip1-105 Aip1-107, Aip1-108, Aip1-109, and Aip1-119. Error bars, SD. The dotted line separates effects of Aip1 proteins with pseudowild-type phenotypes in vivo (white columns) versus those with moderate or severe (light and dark gray columns, respectively) phenotypes in vivo (see Table 1).
Figure 6.
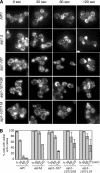
Defects in actin patch turnover in aip1 mutant cells. (A) Wild-type (AIP1) and mutant (aip1Δ, aip1-107, aip1-107/108, aip1-107/119) yeast cells were treated with 50 μM latrunculin A (Lat-A). Samples of cells were removed at the indicated time points, fixed, and stained with Alexa-488 phalloidin. (B) Cells from A were scored for visible actin patches and graphed. More than 200 cells were counted for each time point; columns are the average of two independent experiments. Error bars, SD.
Figure 7.
Defects in actin cable turnover in aip1 mutant cells. (A) Wild-type (AIP1) and mutant (aip1Δ, aip1-107, aip1-107/108, aip1-107/119) yeast cells were treated with 20 μM latrunculin A (Lat-A). Samples of cells were removed at the indicated time points, fixed, and stained with anti-actin antibodies. (B) Cells from A were scored for visible actin cables and graphed. More than 200 cells were counted for each time point, and the columns were the average of two independent experiments. Defects in cable turnover were obvious in aip1 mutant cells at all stages of the cell cycle (unpublished data). These defects were quantified in small- and medium-budded cells, where cables were more prominent and visible than in larger budded cells. Error bars, SD.
Figure 8.
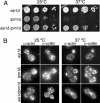
(A) Haploid strains were grown to log phase (OD600 = 0.5) and then cells were serially diluted, plated on YPD medium, and grown for 2 d at 25 or 37°C. (B) Cells were fixed and labeled with rabbit anti-actin and chicken anti-cofilin antibodies after growth at 25°C (left panel) or 37°C (right panel). Note the obvious loss of actin cables in tpm1Δ mutants and their restoration in aip1Δ tpm1Δ mutants.
Figure 9.
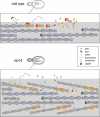
A model for cofilin and Aip1 cellular function: coordinated pruning of tropomyosin-decorated actin cables. In this model, cofilin binds cooperatively to actin filaments in cables, helping to displace tropomyosin from a subset of filaments. The fate of cofilin-severed filaments differs greatly depending on whether Aip1 is available. In wild-type cells, Aip1 may assist cofilin in severing filaments and then rapidly caps the new barbed ends of filaments generated by severing. This leads to rapid net disassembly of those short filaments from their pointed ends. Thus cofilin-decorated filaments in actin cables are extremely short-lived and thus not easily detected in wild-type cells. However, in aip1Δ cells, cofilin severing generates uncapped barbed ends of filaments, which either reanneal or undergo rapid growth, leading to cable thickening. Further, in aip1Δ cells, cofilin-decorated of these cables can be detected by immunostaining because the filaments do not turnover as rapidly as in wild-type cells.
Similar articles
- Intrinsic capability of budding yeast cofilin to promote turnover of tropomyosin-bound actin filaments.
Fan X, Martin-Brown S, Florens L, Li R. Fan X, et al. PLoS One. 2008;3(11):e3641. doi: 10.1371/journal.pone.0003641. Epub 2008 Nov 4. PLoS One. 2008. PMID: 18982060 Free PMC article. - Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1.
Balcer HI, Goodman AL, Rodal AA, Smith E, Kugler J, Heuser JE, Goode BL. Balcer HI, et al. Curr Biol. 2003 Dec 16;13(24):2159-69. doi: 10.1016/j.cub.2003.11.051. Curr Biol. 2003. PMID: 14680631 - A genetic dissection of Aip1p's interactions leads to a model for Aip1p-cofilin cooperative activities.
Clark MG, Teply J, Haarer BK, Viggiano SC, Sept D, Amberg DC. Clark MG, et al. Mol Biol Cell. 2006 Apr;17(4):1971-84. doi: 10.1091/mbc.e05-10-0956. Epub 2006 Jan 18. Mol Biol Cell. 2006. PMID: 16421248 Free PMC article. - Functions of actin-interacting protein 1 (AIP1)/WD repeat protein 1 (WDR1) in actin filament dynamics and cytoskeletal regulation.
Ono S. Ono S. Biochem Biophys Res Commun. 2018 Nov 25;506(2):315-322. doi: 10.1016/j.bbrc.2017.10.096. Epub 2017 Oct 19. Biochem Biophys Res Commun. 2018. PMID: 29056508 Free PMC article. Review.
Cited by
- Actin and endocytosis in budding yeast.
Goode BL, Eskin JA, Wendland B. Goode BL, et al. Genetics. 2015 Feb;199(2):315-58. doi: 10.1534/genetics.112.145540. Genetics. 2015. PMID: 25657349 Free PMC article. Review. - Single-pericyte nanomechanics measured by contraction cytometry.
Islam MM, Gaska I, Oshinowo O, Otumala A, Shekhar S, Au Yong N, Myers DR. Islam MM, et al. APL Bioeng. 2024 Aug 9;8(3):036109. doi: 10.1063/5.0213761. eCollection 2024 Sep. APL Bioeng. 2024. PMID: 39131206 Free PMC article. - Actin dynamics and endocytosis in yeast and mammals.
Galletta BJ, Mooren OL, Cooper JA. Galletta BJ, et al. Curr Opin Biotechnol. 2010 Oct;21(5):604-10. doi: 10.1016/j.copbio.2010.06.006. Epub 2010 Jul 14. Curr Opin Biotechnol. 2010. PMID: 20637595 Free PMC article. Review. - Reconstitution and dissection of the 600-kDa Srv2/CAP complex: roles for oligomerization and cofilin-actin binding in driving actin turnover.
Quintero-Monzon O, Jonasson EM, Bertling E, Talarico L, Chaudhry F, Sihvo M, Lappalainen P, Goode BL. Quintero-Monzon O, et al. J Biol Chem. 2009 Apr 17;284(16):10923-34. doi: 10.1074/jbc.M808760200. Epub 2009 Feb 6. J Biol Chem. 2009. PMID: 19201756 Free PMC article.
References
- Aizawa H., Katadae M., Maruya M., Sameshima M., Murakami-Murofushi K., Yahara I. Hyperosmotic stress-induced reorganization of actin bundles in Dictyostelium cells over-expressing cofilin. Genes Cells. 1999;4:311–324. - PubMed
- Amatruda J. F., Cannon J. F., Tatchell K., Hug C., Cooper J. A. Disruption of the actin cytoskeleton in yeast capping protein mutants. Nature. 1990;344:352–354. - PubMed
- Amberg D. C., Basart E., Botstein D. Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 1995;2:28–35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous