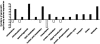Loss of TSLC1 causes male infertility due to a defect at the spermatid stage of spermatogenesis - PubMed (original) (raw)
Loss of TSLC1 causes male infertility due to a defect at the spermatid stage of spermatogenesis
Louise van der Weyden et al. Mol Cell Biol. 2006 May.
Abstract
Tumor suppressor of lung cancer 1 (TSLC1), also known as SgIGSF, IGSF4, and SynCAM, is strongly expressed in spermatogenic cells undergoing the early and late phases of spermatogenesis (spermatogonia to zygotene spermatocytes and elongating spermatids to spermiation). Using embryonic stem cell technology to generate a null mutation of Tslc1 in mice, we found that Tslc1 null male mice were infertile. Tslc1 null adult testes showed that spermatogenesis had arrested at the spermatid stage, with degenerating and apoptotic spermatids sloughing off into the lumen. In adult mice, Tslc1 null round spermatids showed evidence of normal differentiation (an acrosomal cap and F-actin polarization indistinguishable from that of wild-type spermatids); however, the surviving spermatozoa were immature, malformed, found at very low levels in the epididymis, and rarely motile. Analysis of the first wave of spermatogenesis in Tslc1 null mice showed a delay in maturation by day 22 and degeneration of round spermatids by day 28. Expression profiling of the testes revealed that Tslc1 null mice showed increases in the expression levels of genes involved in apoptosis, adhesion, and the cytoskeleton. Taken together, these data show that Tslc1 is essential for normal spermatogenesis in mice.
Figures
FIG. 1.
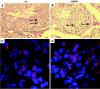
Generation of null and conditional alleles of Tslc1. (A) Tumor suppressor of lung cancer 1 (Tslc1) is a 10-exon gene. loxP (L) sites were positioned to allow Cre-mediated excision of the transmembrane domain (exon 9) to generate a knockout (null) allele. The loxP_-flanked genomic segment is located between nucleotide positions 47,952,177 and 47,953,164 on chromosome 9 of Ensembl (NCBI Mouse Genome Assembly m33). The puroΔtk cassette, flanked by FRT (F) sites, can be excised by using Flp to generate a conditional allele. Southern blot analysis of the Tslc1 targeted (m1), null (m2), and conditional alleles (c1) was performed using two probes, pAI3 and pAJ10, which are 5′ and 3′ of the targeted allele, respectively. Restriction enzyme sites and fragment sizes are indicated. E, EcoRV; P, PacI; S, StuI. (B) Genotyping of Tslc1 null and conditional tail DNAs was performed by PCR using a combination of two primer pairs (either TSL-5/TSL-3 or TSL-C/TSL-3) which can distinguish between all five genotypes (+/+_, +/c1, c1/c1, +/m2, and m2/m2) based on the presence and size of the product(s), which can be resolved in a 2% agarose gel. (C) RT-PCR was performed to confirm deletion of the _loxP_-flanked exons in Tslc1 null mice. RT-PCR performed on RNAs extracted from wild-type mice showed a 245-bp product corresponding to Tslc1 exons 9 and 10. In contrast, no such product was seen for RNAs from m2/m2 mice. RNA from wild-type mice showed 291- and 207-bp products corresponding to exons 7 to 10 and 8 to 10 (reflecting the alternative isoform of murine Tslc1 that differs by the 84 bp that constitute exon 8), whereas RNA from m2/m2 mice produced 159- and 75-bp products, reflecting the deletion of exon 9. β-actin was used as a positive control for cDNA production. (D) Immunohistochemistry performed on testis sections from 3-month-old wild-type and m2/m2 mice, using antibodies against the extracellular domain (EC2) and the C-terminal tail (CC2) of TSLC1.
FIG. 2.
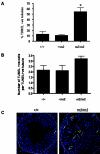
Analysis of wild-type and Tslc1 null testes and epididymides. Hematoxylin- and eosin-stained sections of (A) wild-type testis, (B) Tslc1 null testis, (C) wild-type epididymis, (D) Tslc1 null epididymis, and (E) Tslc1 null testis from mice at 3 months of age are shown. Hematoxylin- and eosin-stained testis sections from (F) Tslc1 null and (G) wild-type mice at 5 months of age are also shown. Magnification, ×400 for panels A to D, ×100 for panels E to G, and ×400 for the inset of panel E. Abbreviations: LC, Leydig cells; Sg, spermatogonia; Sc, spermatocytes; St, spermatids (round and elongated); Sz, spermatozoa; iSz, immature spermatozoa; dSt, degenerated spermatids. (H) DNA flow cytometry analysis of testicular cell suspensions from wild-type and Tslc1 null (m2/m2) mice at 3 months of age (n = 4 or 5 for each genotype). Arrows highlight the differences between the wild-type and Tslc1 null samples. Abbreviations: HC, elongated spermatids; 1C, round spermatids; 2C, spermatogonia; S-ph, spermatogonia synthesizing DNA; 4C, pachytene spermatocytes and G2 spermatogonia. (I) Real-time quantitative RT-PCR of three spermatid genes, Cklf1, Xmr, and Ssty, on RNAs extracted from the testes of Tslc1 null mice at 3 months of age. The results were normalized to β-actin and shown as relative changes in expression level compared with the wild type. Asterisks indicate statistical significance in the relative changes in transcript levels between the wild-type and m2/m2 samples (P < 0.05).
FIG.3.
Tslc1 null testes show increased numbers of apoptotic cells. Tslc1 null (m2/m2) mice show a statistically significant increase in the percentage of TUNEL-positive tubules compared with wild-type (+/+) mice (as indicated by the asterisk; P < 0.05) (A) but not in the number of TUNEL-positive cells per TUNEL-positive tubule (B). (C) Detection of apoptosis (TUNEL-positive cells are indicated by arrows) in testis sections from wild-type mice at 3 months of age, with apoptosis occurring in the spermatogonia, in contrast to Tslc1 null mice, which show apoptosis occurring in the spermatids as well as the spermatogonia. Slides for three mice of each genotype were examined, and representative slides are shown. Magnification, ×480.
FIG. 4.
Delayed and disrupted first wave of spermatogenesis in Tslc1 null mice. Hematoxylin- and eosin-stained sections of testes from wild-type (+/+) and Tslc1 null (m2/m2) mice at postnatal days 7, 14, 22, 28, and 35 are shown. Magnification for first and second columns, ×200 for day 7 images and ×400 for day 14 to 35 images; magnification for third column, ×1,000. Abbreviations: Sg, spermatogonia; 1° Sp, primary spermatocytes; 2° Sp, secondary spermatocytes; St, spermatids (round and elongated); dSt, degenerated spermatids; Sz, spermatozoa; iSz, immature spermatozoa; MNC, multinucleate giant cells (seen in m2/m2 mice on days 22, 28, and 35; shown at a ×400 magnification).
FIG. 5.
Tslc1 null testes show some features of differentiation. Histochemical analysis of testis sections from (A) wild-type and (B) Tslc1 null (m2/m2) mice by periodic acid and Schiff staining showed the presence of acrosomal caps (stained pink) in the spermatids (St) and spermatozoa (Sz). Slides for three mice of each genotype were examined, and representative images are shown. Magnification, ×400. An analysis of testis sections from (C) wild-type and (D) Tslc1 null (m2/m2) mice by phalloidin staining showed normal F-actin (red) polarization in the spermatids of both. Slides for three mice of each genotype were examined, and representative images are shown. Magnification, ×1,600.
FIG. 6.
Analysis of wild-type and Tslc1 null spermatozoa. Phase-contrast light microscopy showed that in contrast to wild-type (A) or Tslc1 heterozygous (B) spermatozoa, Tslc1 null spermatozoa showed morphological abnormalities, as indicated by the arrows (C to J). Images shown are representative of spermatozoa from the caudal epididymides of three to five mice of each genotype. Magnification, ×400. Electron microscopy analysis showed normal sperm head nuclei from wild-type mice (K), in contrast to sperm heads from Tslc1 null mice, which showed abnormally shaped nuclei (L) and enlarged nuclei with abnormal residual cytoplasm, often showing residual body-like changes (M and O), degenerated nuclei (N), dilated mitochondria and vacuoles (P), and multiple malformed nuclei within a single cellular cytoplasm (Q and R). Abbreviations: N, nucleus; C, cytoplasm; RB, residual body-like changes. Sections from two mice of each genotype were examined, and representative images are shown. Magnification, ×4,500 for panels K to N, Q, and R and ×10,000 for panels O and P.
FIG. 7.
Microarray analysis of testis RNAs from Tslc1 null mice. Germ cell-specific microarray analysis was used to identify differentially expressed genes in the testes of Tslc1 null mice compared with those of wild-type mice. The data set was normalized and filtered to select differentially expressed genes, with 136 genes being selected as significantly altered in expression in Tslc1 null testes relative to wild-type testes (see Materials and Methods). There were many more upregulated genes (103/136) than downregulated genes, and the differentially expressed genes are shown by classification according to their likely biological function in Onto-Express.
Similar articles
- Disruption of spermatogenic cell adhesion and male infertility in mice lacking TSLC1/IGSF4, an immunoglobulin superfamily cell adhesion molecule.
Yamada D, Yoshida M, Williams YN, Fukami T, Kikuchi S, Masuda M, Maruyama T, Ohta T, Nakae D, Maekawa A, Kitamura T, Murakami Y. Yamada D, et al. Mol Cell Biol. 2006 May;26(9):3610-24. doi: 10.1128/MCB.26.9.3610-3624.2006. Mol Cell Biol. 2006. PMID: 16612000 Free PMC article. - Oligo-astheno-teratozoospermia in mice lacking RA175/TSLC1/SynCAM/IGSF4A, a cell adhesion molecule in the immunoglobulin superfamily.
Fujita E, Kouroku Y, Ozeki S, Tanabe Y, Toyama Y, Maekawa M, Kojima N, Senoo H, Toshimori K, Momoi T. Fujita E, et al. Mol Cell Biol. 2006 Jan;26(2):718-26. doi: 10.1128/MCB.26.2.718-726.2006. Mol Cell Biol. 2006. PMID: 16382161 Free PMC article. - Loss of partitioning-defective-3/isotype-specific interacting protein (par-3/ASIP) in the elongating spermatid of RA175 (IGSF4A/SynCAM)-deficient mice.
Fujita E, Tanabe Y, Hirose T, Aurrand-Lions M, Kasahara T, Imhof BA, Ohno S, Momoi T. Fujita E, et al. Am J Pathol. 2007 Dec;171(6):1800-10. doi: 10.2353/ajpath.2007.070261. Epub 2007 Nov 30. Am J Pathol. 2007. PMID: 18055550 Free PMC article. - IGSF4: a new intercellular adhesion molecule that is called by three names, TSLC1, SgIGSF and SynCAM, by virtue of its diverse function.
Watabe K, Ito A, Koma YI, Kitamura Y. Watabe K, et al. Histol Histopathol. 2003 Oct;18(4):1321-9. doi: 10.14670/HH-18.1321. Histol Histopathol. 2003. PMID: 12973698 Review. - Involvement of a cell adhesion molecule, TSLC1/IGSF4, in human oncogenesis.
Murakami Y. Murakami Y. Cancer Sci. 2005 Sep;96(9):543-52. doi: 10.1111/j.1349-7006.2005.00089.x. Cancer Sci. 2005. PMID: 16128739 Free PMC article. Review.
Cited by
- CRTAM controls residency of gut CD4+CD8+ T cells in the steady state and maintenance of gut CD4+ Th17 during parasitic infection.
Cortez VS, Cervantes-Barragan L, Song C, Gilfillan S, McDonald KG, Tussiwand R, Edelson BT, Murakami Y, Murphy KM, Newberry RD, Sibley LD, Colonna M. Cortez VS, et al. J Exp Med. 2014 Apr 7;211(4):623-33. doi: 10.1084/jem.20130904. Epub 2014 Mar 31. J Exp Med. 2014. PMID: 24687959 Free PMC article. - Histopathologic effects of mobile phone radiation exposure on the testes and sperm parameters: a systematic literature review of animal studies.
Assefa EM, Abdu SM. Assefa EM, et al. Front Reprod Health. 2025 Jan 17;6:1515166. doi: 10.3389/frph.2024.1515166. eCollection 2024. Front Reprod Health. 2025. PMID: 39896841 Free PMC article. - Cell adhesion, the backbone of the synapse: "vertebrate" and "invertebrate" perspectives.
Giagtzoglou N, Ly CV, Bellen HJ. Giagtzoglou N, et al. Cold Spring Harb Perspect Biol. 2009 Oct;1(4):a003079. doi: 10.1101/cshperspect.a003079. Cold Spring Harb Perspect Biol. 2009. PMID: 20066100 Free PMC article. Review. - Rapid assembly of functional presynaptic boutons triggered by adhesive contacts.
Lucido AL, Suarez Sanchez F, Thostrup P, Kwiatkowski AV, Leal-Ortiz S, Gopalakrishnan G, Liazoghli D, Belkaid W, Lennox RB, Grutter P, Garner CC, Colman DR. Lucido AL, et al. J Neurosci. 2009 Oct 7;29(40):12449-66. doi: 10.1523/JNEUROSCI.1381-09.2009. J Neurosci. 2009. PMID: 19812321 Free PMC article. - Silencing of ErbB3/ErbB2 signaling by immunoglobulin-like Necl-2.
Kawano S, Ikeda W, Kishimoto M, Ogita H, Takai Y. Kawano S, et al. J Biol Chem. 2009 Aug 28;284(35):23793-805. doi: 10.1074/jbc.M109.025155. Epub 2009 Jun 26. J Biol Chem. 2009. PMID: 19561085 Free PMC article.
References
- Biederer, T., Y. Sara, M. Mozhayeva, D. Atasoy, X. Liu, E. T. Kavalali, and T. C. Sudhof. 2002. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 297:1525-1531. - PubMed
- Blanco-Rodriguez, J. 1998. A matter of death and life: the significance of germ cell death during spermatogenesis. Int. J. Androl. 21:236-248. - PubMed
- Blanco-Rodriguez, J., C. Martinez-Garcia, and A. Porras. 2003. Correlation between DNA synthesis in the second, third and fourth generations of spermatogonia and the occurrence of apoptosis in both spermatogonia and spermatocytes. Reproduction 126:661-668. - PubMed
- Boles, K. S., W. Barchet, T. Diacovo, M. Cella, and M. Colonna. 2005. The tumor suppressor TSLC1/NECL-2 triggers NK cell and CD8+ T cell responses through the cell surface receptor CRTAM. Blood 106:779-786. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous