A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities - PubMed (original) (raw)
. 2006 May 1;20(9):1110-22.
doi: 10.1101/gad.377406. Epub 2006 Apr 17.
Affiliations
- PMID: 16618800
- PMCID: PMC1472471
- DOI: 10.1101/gad.377406
A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities
Tetyana Klymenko et al. Genes Dev. 2006.
Abstract
Polycomb response elements (PREs) are specific cis-regulatory sequences needed for transcriptional repression of HOX and other target genes by Polycomb group (PcG) proteins. Among the many PcG proteins known in Drosophila, Pho is the only sequence-specific DNA-binding protein. To gain insight into the function of Pho, we purified Pho protein complexes from Drosophila embryos and found that Pho exists in two distinct protein assemblies: a Pho-dINO80 complex containing the Drosophila INO80 nucleosome-remodeling complex, and a Pho-repressive complex (PhoRC) containing the uncharacterized gene product dSfmbt. Analysis of PhoRC reveals that dSfmbt is a novel PcG protein that is essential for HOX gene repression in Drosophila. PhoRC is bound at HOX gene PREs in vivo, and this targeting strictly depends on Pho-binding sites. Characterization of dSfmbt protein shows that its MBT repeats have unique discriminatory binding activity for methylated lysine residues in histones H3 and H4; the MBT repeats bind mono- and di-methylated H3-K9 and H4-K20 but fail to interact with these residues if they are unmodified or tri-methylated. Our results establish PhoRC as a novel Drosophila PcG protein complex that combines DNA-targeting activity (Pho) with a unique modified histone-binding activity (dSfmbt). We propose that PRE-tethered PhoRC selectively interacts with methylated histones in the chromatin flanking PREs to maintain a Polycomb-repressed chromatin state.
Figures
Figure 1.
TAP of Pho protein complexes from Drosophila embryos. (A) Western blot of total embryo extracts from wild-type (lanes 1,2) or pho1/pho1 mutant embryos (lane 3) that carry the Pho-TAP transgene as indicated, probed with anti-Pho antibody. Note, pho1 is a protein-null allele. Stoichiometry of Pho and Pho-TAP protein cannot be compared since Pho antibody binds to Pho epitope and the protein A moiety in the Pho-TAP protein. (B) Pho protein complexes purified from nuclear extract prepared from wild-type (wt) or Pho-TAP, pho1/pho1 (Pho-TAP) Drosophila embryos visualized by silver staining. Input material for mock-purification from wild-type embryos and for purification from Pho-TAP embryos was normalized by protein concentration. Equivalent amounts of calmodulin-affinity resin was boiled in SDS sample buffer, and eluted material was separated on a 4%–12% polyacrylamide gel. The indicated proteins consistently copurified with Pho-CBP protein in several independent experiments; copurification of these proteins with Pho-CBP was independent of the genetic background (i.e., pho+ or pho1/pho1). (Pho-CBP) Fusion protein containing the calmodulin-binding moiety of the TAP tag. Lower-molecular-weight dSfmbt bands were identified by microsequencing, and they likely correspond to dSfmbt degradation products. Asterisks indicate bands for which we have not been able to obtain unambiguous peptide sequence data; see Supplemental Material S1A for information on additional proteins that were identified by mass spectrometry. (C) Western blot analysis of total embryonic nuclear extract input material (IN, lanes 1,2) from wild-type (wt) and Pho-TAP transgenic embryos and material eluted from calmodulin affinity resin (E, lanes 3,4) after purification. All panels come from the same batch of input material, and the eluates were all from the same batch of material purified from wild-type and Pho-TAP embryos, respectively. Note that the Pho-TAP embryos used for this experiment were pho+, but the same results were obtained if complexes were purified from Pho-TAP, pho1/pho1 embryos. Asterisks indicate endogenous Pho (red), Pho-CBP (blue), and Pho-TAP (green) protein; in lane 2, Pho-TAP is also detected by other antibodies due to protein A tag. Note signals for dSfmbt, Reptin, and Pontin but a lack of signals for E(z), Su(z)12, Pc, and Scm in lane 4. (D) Domain architecture of the three Drosophila MBT repeat proteins dSfmbt, l(3)mbt, and Scm and alignment of the corresponding MBT repeats. The mouse Sfmbt protein (mSfmbt) as described by Usui et al. (2000) is shown for comparison.
Figure 2.
dSfmbt and dINO80 exist in distinct Pho protein complexes. (A) Crude embryonic nuclear extracts were separated on a 20%–50% glycerol gradient. Fractions were probed with antibodies against dINO80, dSfmbt, and Pho; (IN) nuclear extract input material. Thyroglobulin (669 kDa), catalase (232 kDa), and aldolase (158 kDa) were fractionated on a separate gradient that was run in parallel, and fractions containing these proteins are indicated on top. Note that dINO80 and dSfmbt are present in distinct complexes, both of which contain Pho; the presence of Pho in other fractions suggests that Pho may also exist in additional complexes. Pho-like is present across the whole gradient similarly to Pho (not shown). (B) Immunoprecipitations from embryonic nuclear extracts were probed with antibodies indicated on the left. (Lane 1) Nuclear extract input material (IN, 10%) used for immunoprecipitation with preimmune serum (Pre-I, lane 2) or with antibodies against dSfmbt (lane 3) or dINO80 (lane 4). Pho is present in samples immunoprecipitated with dSfmbt or dINO80 antibodies, but dINO80 is not detected in dSfmbt precipitates, and dSfmbt is absent from dINO80 precipitates. Pho-like is enriched in samples immunoprecipitated with dSfmbt, but background bands in dINO80 IP reactions (asterisks) do not permit unambiguous detection of Pho-like protein. Note the absence of Scm, Ph, Pc, or Su(z)12 in both dSfmbt and dINO80 immunoprecipitates; asterisks in the Scm Western blot panel indicate nonspecific bands.
Figure 3.
Binding of dSfmbt to PREs depends on Pho protein-binding sites. (Top) X-ChIP analysis on PRED transformant embryos. Pho, dSfmbt, and Ph are specifically bound at the bxd and iab-7 PREs in the Ubx and Abd-B loci, respectively, and to the wild-type bxd PRE in the PRED reporter gene (Fritsch et al. 1999). The location of PREs with respect to the Ubx or Abd-B transcription start site is indicated in kilobases. No binding is detected in regions flanking the PREs and at two intergenic locations elsewhere in the genome. The embryos carried a single copy of the PRED transgene. (Bottom) X-ChIP analysis on PRED Pho mut transformant embryos. Binding of Pho, dSfmbt, and Ph at PRED Pho mut is strongly reduced due to mutation of all six Pho protein-binding sites in the PRED Pho mut fragment (Fritsch et al. 1999). Binding signals at the endogenous bxd and iab-7 PREs is comparable in chromatin from PRED and from PRED Pho mut embryos. X-ChIP signals are represented as the fraction (%) of input material precipitated in each IP reaction; signals represent results from immunoprecipitation reactions performed on three independently purified batches of chromatin.
Figure 4.
Reconstitution of recombinant PhoRC. (Layout in A_–_C, top) Indicated Flag-tagged (red +) and untagged (black +) proteins were (co)expressed in Sf9 cells, affinity-purified via the Flag tag, separated by SDS-PAGE, and visualized by coomassie staining. (Below) Western blot of corresponding Sf9 total cell extracts prior to purification to reveal coexpression of proteins at comparable levels. (A) dSfmbt and Pho form a stable complex. (Top) Purified PhoRC contains comparable amounts of dSfmbt and Flag-Pho proteins (lane 3), but Sfmbt and Pho do not copurify with Flag-Reptin, Flag-Pontin, Flag-Esc, or Flag-Pc, respectively (lanes 4_–_9). (Lanes 6,7) As yeast Rvb1p/Rvb2p (Jonsson et al. 2004), Reptin and Pontin are expected to form a stable complex, and the presence of this complex thus provides an internal control for complex reconstitution. (Below) Western signals show that dSfmbt and Flag-tagged proteins were coexpressed at similar ratios in all experiments. (B, lane 3) dSfmbt and Pho-like form a stable complex that contains comparable amounts of both proteins. (C) Pho does not form stable complexes with Esc and/or E(z). Note that Pho does not copurify either with Flag-Esc (lane 2) or with Flag-E(z) (lane 4) alone and that stoichiometric Flag-Esc–E(z) complexes without Pho are purified if Flag-Esc, E(z), and Pho are coexpressed (lane 3).
Figure 5.
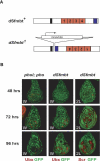
dSfmbt is a novel PcG gene required for HOX gene silencing. (A) Structure of the dSfmbt1 knockout allele generated by homologous recombination. In the disruption allele, the whole 3′ coding region was inserted in the inverted orientation (see Material and Methods). (B) Imaginal discs with clones of cells that are homozygous for dSfmbt1 (dSfmbt) or double homozygous for pho-like81A and pho1 (phol; pho) were stained with antibodies against Ubx or Scr (red) and GFP (green); the clones of dSfmbt single-mutant or phol; pho double-mutant cells are marked by the absence of GFP. Discs were analyzed 48, 72, or 96 h after clone induction as indicated. (Middle column) Widespread misexpression of Ubx is seen in dSfmbt mutant clones in the wing (W) disc 72 and 96 h after clone induction; Ubx is normally not expressed in the wing disc. As in the case of other PcG mutants (Beuchle et al. 2001), Ubx is still silenced in some Sfmbt mutant clones (i.e., in the notum), even 96 h after clone induction. (Right column) Widespread misexpression of Scr is seen in dSfmbt mutant clones in the second leg (2L) disc 72 and 96 h after clone induction; Scr is normally not expressed in the second leg disc. (Left column) In phol; pho double-mutant clones, misexpression of Ubx is seen 48 and 72 h after clone induction. Ninety-six hours after clone induction, most clones of phol; pho double-mutant cells have been eliminated from the disc; note the lack of GFP-negative clones compared with the brightly labeled GFP+/GFP+ twin spot clones that were induced by the reciprocal recombination event at the time of clone induction. 2L disc images were enlarged; 2L discs are in reality half the size of W discs.
Figure 6.
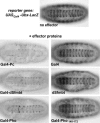
DNA-tethered dSfmbt represses transcription. Ventral views of 14–16-h-old transformant embryos that carry one copy of the UASGal4-Ubx-LacZ reporter gene (BGUZ construct) (Müller 1995) and one copy of the indicated hsp70-effector construct (“Gal4” corresponds to Gal41–147, the DNA-binding domain of Gal4); in all cases, embryos were heat-shocked (see Materials and Methods) and stained with antibody against β-gal. Repression by Gal4-dSfmbt and Gal4-Pho is almost as effective as repression caused by Gal4-Pc protein (Müller 1995). Expression of the Gal4 DNA-binding domain alone or dSfmbt alone does not result in detectable repression. Gal-Pho140–172 is also not effective as a repressor in this assay. Pho140–172 includes the so-called spacer domain (Brown et al. 1998) that has been implicated in physical interactions with PRC1 components Pc and Ph (Mohd-Sarip et al. 2002).
Figure 7.
Selective binding of dSfmbt MBT repeats to histone H3 and H4 tail peptides that carry specific methyl-lysine modifications. (A) Peptide backbones corresponding to different regions of the H3 and H4 N-terminal tail used in interaction studies. Differentially methylated lysine residues are indicated in red. (B) Binding of the MBT repeats of dSfmbt to H3 peptides that are differentially methylated on K9 and to H4 peptides differentially methylated on K20 as measured by fluorescence anisotropy. Data points correspond to averages from three independent experiments. (C) Dissociation constants (Kd in micromolar, μM) of the interaction of the MBT repeats of dSfmbt with different modified H3 and H4 peptides. Values correspond to averages from at least three independent experiments.
Similar articles
- Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt.
Grimm C, Matos R, Ly-Hartig N, Steuerwald U, Lindner D, Rybin V, Müller J, Müller CW. Grimm C, et al. EMBO J. 2009 Jul 8;28(13):1965-77. doi: 10.1038/emboj.2009.147. Epub 2009 Jun 4. EMBO J. 2009. PMID: 19494831 Free PMC article. - Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins.
Papp B, Müller J. Papp B, et al. Genes Dev. 2006 Aug 1;20(15):2041-54. doi: 10.1101/gad.388706. Genes Dev. 2006. PMID: 16882982 Free PMC article. - The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing.
Brown JL, Fritsch C, Mueller J, Kassis JA. Brown JL, et al. Development. 2003 Jan;130(2):285-94. doi: 10.1242/dev.00204. Development. 2003. PMID: 12466196 - Polycomb response elements and targeting of Polycomb group proteins in Drosophila.
Müller J, Kassis JA. Müller J, et al. Curr Opin Genet Dev. 2006 Oct;16(5):476-84. doi: 10.1016/j.gde.2006.08.005. Epub 2006 Aug 17. Curr Opin Genet Dev. 2006. PMID: 16914306 Review. - Chromatin regulation: how complex does it get?
Meier K, Brehm A. Meier K, et al. Epigenetics. 2014 Nov;9(11):1485-95. doi: 10.4161/15592294.2014.971580. Epigenetics. 2014. PMID: 25482055 Free PMC article. Review.
Cited by
- Pleiohomeotic interacts with the core transcription elongation factor Spt5 to regulate gene expression in Drosophila.
Harvey R, Schuster E, Jennings BH. Harvey R, et al. PLoS One. 2013 Jul 22;8(7):e70184. doi: 10.1371/journal.pone.0070184. Print 2013. PLoS One. 2013. PMID: 23894613 Free PMC article. - Gene silencing and Polycomb group proteins: an overview of their structure, mechanisms and phylogenetics.
Golbabapour S, Majid NA, Hassandarvish P, Hajrezaie M, Abdulla MA, Hadi AH. Golbabapour S, et al. OMICS. 2013 Jun;17(6):283-96. doi: 10.1089/omi.2012.0105. OMICS. 2013. PMID: 23692361 Free PMC article. Review. - DNA sequence models of genome-wide Drosophila melanogaster Polycomb binding sites improve generalization to independent Polycomb Response Elements.
Bredesen BA, Rehmsmeier M. Bredesen BA, et al. Nucleic Acids Res. 2019 Sep 5;47(15):7781-7797. doi: 10.1093/nar/gkz617. Nucleic Acids Res. 2019. PMID: 31340029 Free PMC article. - Identification of hepatocellular carcinoma subtypes based on PcG-related genes and biological relevance with cancer cells.
Fu Y, Yang K, Wu K, Wang H, Li Q, Zhang F, Yang K, Yao Q, Ma X, Deng Y, Zhang J, Liu C, Qu K. Fu Y, et al. Clin Epigenetics. 2022 Dec 24;14(1):184. doi: 10.1186/s13148-022-01393-6. Clin Epigenetics. 2022. PMID: 36566204 Free PMC article. - Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes.
Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Köcher T, Cohen A, Stunnenberg HG, Wilm M, Müller J. Nekrasov M, et al. EMBO J. 2007 Sep 19;26(18):4078-88. doi: 10.1038/sj.emboj.7601837. Epub 2007 Aug 30. EMBO J. 2007. PMID: 17762866 Free PMC article.
References
- Beuchle D., Struhl G., Muller J. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development. 2001;128:993–1004. - PubMed
- Birve A., Sengupta A.K., Beuchle D., Larsson J., Kennison J.A., Rasmuson-Lestander A., Müller J. Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development. 2001;128:3371–3379. - PubMed
- Bonaldi T., Imhof A., Regula J.T. A combination of different mass spectroscopic techniques for the analysis of dynamic changes of histone modifications. Proteomics. 2004;4:1382–1396. - PubMed
- Breen T.R., Duncan I.M. Maternal expression of genes that regulate the bithorax complex of. Drosophila melanogaster. Dev. Biol. 1986;118:442–456. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous