Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity - PubMed (original) (raw)
Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity
Maria del Carmen Seleme et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Despite being scarce in the human genome, active L1 retrotransposons continue to play a significant role in its evolution. Because of their recent expansion, many L1s are not fixed in humans, and, when present, their mobilization potential can vary among individuals. Previously, we showed that the great majority of retrotransposition events in humans are caused by highly active, or hot, L1s. Here, in four populations of diverse geographic origins (160 haploid genomes), we investigated the degree of sequence polymorphism of three hot L1s and the extent of individual variation in mobilization capability of their allelic variants. For each locus, we found one previously uncharacterized allele in every three to five genomes, including some with nonsense and insertion/deletion mutations. Single or multiple nucleotide substitutions drastically affected the retrotransposition efficiency of some alleles. One-third of elements were no longer hot, and these so-called cool alleles substantially increased the range of individual susceptibility to retrotransposition events. Adding the activity of the three elements in each individual resulted in a surprising degree of variation in mobilization capability, ranging from 0% to 390% of a reference L1. These data suggest that individual variation in retrotransposition potential makes an important contribution to human genetic diversity.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Fig. 1.
Alleles and activity variants of L1A, L1B, and L1C. (A_–_C Top) Nucleotide changes relative to HGWD sequence (allele 1). Amino acid changes are in parentheses. The retrotransposition activity (%L1rp) of each allele is shown at left. (A_–_C Bottom) A scaled L1 sequence. Lines indicate the location of each change. ORF1 and ORF2 (gray boxes) appear separated by the inter ORF (white box). Hatched boxes represent (left to right) leucine zipper, endonuclease, reverse transcriptase, and zinc knuckle. Black boxes in ORF2 represent sites A and B, putative ORF1p-binding sites to L1 RNA (57). (A) L1A. 17 polymorphic sites distributed in 8 alleles (35 genomes). A circle denotes the change responsible for an 87% reduction in activity. (B) L1B. Nineteen polymorphic sites distributed in 18 alleles (59 genomes). Circles indicate potential changes that reduce activity by 50–88%. (C) L1C. Twenty-six polymorphic sites distributed in 26 alleles (72 genomes). Circles denote changes potentially responsible for an 80% reduction in activity. For alleles marked with an asterisk or denoted nd, the activity was not tested because the alleles could not be cloned. ∗, the activity value was predicted from sequence similarities to closely related, tested alleles; nd, the activity value could not be predicted because the amino acid changes were not present in other alleles.
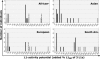
Fig. 2.
Combined retrotransposition potential of three hot L1s per individual in four populations. From 26% (African) to 55% (South American) of individuals per population have a unique L1 activity potential. White, black, and hatched bars represent individuals lacking a hot L1 phenotype (<25%), having an intermediate L1 activity, and having a high L1 activity (>200%), respectively. ∗, The African distribution is based on 19 individuals (Table 5).
Fig. 3.
Average retrotransposition potential of three hot L1s in four populations. The total retrotransposition potential of L1A, L1B, and L1C for each individual was divided by the number of individuals in the population to determine the average retrotransposition potential in each population. The means of the four populations are not equal by ANOVA (P = 0.036).
Fig. 4.
Model of the evolution of an L1 insertion in a population. Data presented here and evidence that hot L1s account for most new insertions (34) suggest that new insertions are derived from hot L1s. Data on alleles of L1A, L1B, L1C, and LRE1 (36, 37) indicate that, after a hot L1 reaches an intermediate gene frequency in the population, it has a significant proportion of cool alleles. As an L1 approaches fixation, mutations produce cool alleles and dead alleles. Shaded box, L1 insertion in chromosomes (lines); black dots, mutations.
Similar articles
- Hot L1s account for the bulk of retrotransposition in the human population.
Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH Jr. Brouha B, et al. Proc Natl Acad Sci U S A. 2003 Apr 29;100(9):5280-5. doi: 10.1073/pnas.0831042100. Epub 2003 Apr 7. Proc Natl Acad Sci U S A. 2003. PMID: 12682288 Free PMC article. - LINE-1 retrotransposition activity in human genomes.
Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. Beck CR, et al. Cell. 2010 Jun 25;141(7):1159-70. doi: 10.1016/j.cell.2010.05.021. Cell. 2010. PMID: 20602998 Free PMC article. - Allelic heterogeneity in LINE-1 retrotransposition activity.
Lutz SM, Vincent BJ, Kazazian HH Jr, Batzer MA, Moran JV. Lutz SM, et al. Am J Hum Genet. 2003 Dec;73(6):1431-7. doi: 10.1086/379744. Epub 2003 Nov 7. Am J Hum Genet. 2003. PMID: 14610717 Free PMC article. - L1 retrotransposons and somatic mosaicism in the brain.
Richardson SR, Morell S, Faulkner GJ. Richardson SR, et al. Annu Rev Genet. 2014;48:1-27. doi: 10.1146/annurev-genet-120213-092412. Epub 2014 Jul 14. Annu Rev Genet. 2014. PMID: 25036377 Review.
Cited by
- Evolutionary insights from profiling LINE-1 activity at allelic resolution in a single human genome.
Yang L, Metzger GA, Padilla Del Valle R, Delgadillo Rubalcaba D, McLaughlin RN Jr. Yang L, et al. EMBO J. 2024 Jan;43(1):112-131. doi: 10.1038/s44318-023-00007-y. Epub 2023 Dec 18. EMBO J. 2024. PMID: 38177314 Free PMC article. - Emerging Opportunities to Study Mobile Element Insertions and Their Source Elements in an Expanding Universe of Sequenced Human Genomes.
Devine SE. Devine SE. Genes (Basel). 2023 Oct 10;14(10):1923. doi: 10.3390/genes14101923. Genes (Basel). 2023. PMID: 37895272 Free PMC article. Review. - Retrotransposon insertions associated with risk of neurologic and psychiatric diseases.
Ahn HW, Worman ZF, Lechsinska A, Payer LM, Wang T, Malik N, Li W, Burns KH, Nath A, Levin HL. Ahn HW, et al. EMBO Rep. 2023 Jan 9;24(1):e55197. doi: 10.15252/embr.202255197. Epub 2022 Nov 11. EMBO Rep. 2023. PMID: 36367221 Free PMC article. - Intragenic L1 Insertion: One Possibility of Brain Disorder.
Son JH, Do H, Han J. Son JH, et al. Life (Basel). 2022 Sep 13;12(9):1425. doi: 10.3390/life12091425. Life (Basel). 2022. PMID: 36143463 Free PMC article. Review. - Somatic retrotransposition in the developing rhesus macaque brain.
Billon V, Sanchez-Luque FJ, Rasmussen J, Bodea GO, Gerhardt DJ, Gerdes P, Cheetham SW, Schauer SN, Ajjikuttira P, Meyer TJ, Layman CE, Nevonen KA, Jansz N, Garcia-Perez JL, Richardson SR, Ewing AD, Carbone L, Faulkner GJ. Billon V, et al. Genome Res. 2022 Jul;32(7):1298-1314. doi: 10.1101/gr.276451.121. Epub 2022 Jun 21. Genome Res. 2022. PMID: 35728967 Free PMC article.
References
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. Nature. 2001;409:860–921. - PubMed
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Science. 2004;305:525–528. - PubMed
- Kazazian H. H., Jr Science. 2004;303:1626–1632. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources