Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations - PubMed (original) (raw)
Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations
Miguel R Branco et al. PLoS Biol. 2006 May.
Abstract
After mitosis, mammalian chromosomes partially decondense to occupy distinct territories in the cell nucleus. Current models propose that territories are separated by an interchromatin domain, rich in soluble nuclear machinery, where only rare interchromosomal interactions can occur via extended chromatin loops. In contrast, recent evidence for chromatin mobility and high frequency of chromosome translocations are consistent with significant levels of chromosome intermingling, with important consequences for genome function and stability. Here we use a novel high-resolution in situ hybridization procedure that preserves chromatin nanostructure to show that chromosome territories intermingle significantly in the nucleus of human cells. The degree of intermingling between specific chromosome pairs in human lymphocytes correlates with the frequency of chromosome translocations in the same cell type, implying that double-strand breaks formed within areas of intermingling are more likely to participate in interchromosomal rearrangements. The presence of transcription factories in regions of intermingling and the effect of transcription impairment on the interactions between chromosomes shows that transcription-dependent interchromosomal associations shape chromosome organization in mammalian cells. These findings suggest that local chromatin conformation and gene transcription influence the extent with which chromosomes interact and affect their overall properties, with direct consequences for cell-type specific genome stability.
Figures
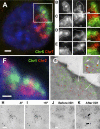
Figure 1. Chromosome Territories Intermingle in the Nucleus of Human Cells
Chromosomes were painted in cryosections (approximately 150 nm thick) of human lymphocytes and visualized by fluorescence microscopy (A–F) or indirectly immunolabeled with gold particles before imaging by electron microscopy (G–I). (A) Example of a nuclear section showing intermingling between Chromosomes 5 and 7. (B–E) Intermingling is best seen in gray-scale images after the mask for one chromosome (white line) is overlaid on the image of the other chromosome. The intersection between masks for both chromosomes is shown in the merged images (yellow line), representing areas of intermingling. Fluorescence intensity line scans of relevant areas in images (B–E) can be in Figure S1. (F and G) Five- and 10-nm gold particles labeling Chromosomes 1 and 2 (G; pseudo-colored green and red, respectively) are intermingled within the intersections identified on the light microscope in the same nuclear section (F). Gold particles labeling different CTs can be found in close proximity within areas of intermingling (G, arrows in inset). (H and I) Stereoviews of a region of colocalization (*) between Chromosome 1 (5 nm gold) and 2 (10 nm gold) were obtained by collecting images tilted −6° (H) and +6° (I) relative to the_z_-axis. 3D visualization shows that differently-sized particles lie adjacent in the same_z_ planes. (J and K) Histone H2B was indirectly immunolabeled with 5-nm gold particles and imaged on the EM before (J) and after (K) mock FISH; gold particles are spatially preserved in both heterochromatic (arrow 1) and euchromatic (arrow 2) regions. Bars: (A and F) 1 μm; (B–E) 0.5 μm; (G–K) 50 nm.
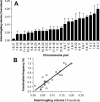
Figure 2. CT Intermingling Correlates with Translocation Potential
Intermingling volumes were measured for 24 pairs of chromosomes in human lymphocytes (A) and plotted against the respective translocation frequency in the same cell type ( Table S1) [ 26] (B), showing a highly significant correlation (p < 0.0001). Error bars represent standard deviations.
Figure 3. Chromosome Intermingling Is Influenced by Transcription-Dependent Interactions
(A–F) Immuno-FISH labeling shows the active form of PolII in regions of intermingling. Serine2-phosphorylated PolII was immunolabeled with H5 antibodies and AlexaFluor488, before hybridization with paints for Chromosome 3 and all other chromosomes (WG-3). A magnified view of Chromosome 3 (A) and the remaining chromatin (B) is shown, which contains a region of intermingling (C). Active PolII is found within masks delineating Chromosome 3 territory (D), the remaining chromatin (E) and regions of intermingling (F). PolII sites are not excluded or enriched in areas of intermingling (see Figure S4). Bar: 1 μm. (G) Measurements of intermingling for ten chromosome pairs in control and α-amanitin–treated lymphocytes reveals changes in intermingling volumes for four of ten pairs analyzed, showing that the extent of intermingling is affected by ongoing transcription. (H) Differences in intermingling are not due to altered CT volumes, as these did not change after treatment with α-amanitin for 12 of 13 chromosomes analyzed. Error bars represent standard deviations (*p < 0.05, **p < 0.01).
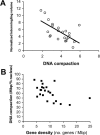
Figure 4. Less Compact Chromosomes Are More Gene-Rich and Tend to Intermingle More
The normalized intermingling volumes for each pair of CTs were plotted against the product of their DNA compaction ratios (A), which were obtained by calculating the respective DNA content per unit of CT volume ( Figure S5). Individual DNA compaction ratios were plotted against the respective gene density (B). The negative correlations observed in both cases reveal that gene-rich chromosomes tend to be less compact, creating a higher potential for nonspecific intermingling. Error bars represent standard deviations.
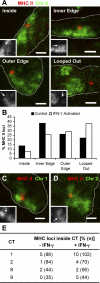
Figure 5. The MHC II Locus Is Found within Other CTs upon IFN-γ Activation
(A and B) A BAC probe for the MHC II locus (red) was cohybridized with a Chromosome 6 paint (green) in cryosections of control and IFN-γ activated MRC5 human lung fibroblasts (nuclear edge outlined by dotted line). Insets show the position of the MRC II locus (arrows) in relation to its CT (in gray scale) (A). The positions of the MRC II loci were scored into four different categories. Loci found “inside” or “looped out” were easily classified; loci near the edge of the CT were divided into “inner edge” and “outer edge,” depending on whether they appeared more internal or external in relation to the remainder of the CT. Upon IFN-γ activation, the MHC II locus relocates to a more external position in relation to its CT, when compared with control cells (B,p = 0.02, two-tailed χ2 test,n = 118 and 117 loci for control and IFN-γ activated cells, respectively), as described before [ 27]. Note that due to the flatness of fibroblast cells, nuclear profiles from random sections are often elongated. (C–E) The MHC II BAC probe (red) was cohybridized with paints for Chromosomes 1 (C, green), 2 (D, green), 8, or 9 (not shown) and the number of MHC II loci found within each of these CTs was scored in both control and IFN-γ activated cells (E). Upon activation, the MHC II locus is more likely to be found within one of Chromosomes 1, 2, or 9, when compared with control cells (p = 0.038, two-tailed Fisher's exact test using pooled data from the three chromosomes), whereas no difference in association is detected if Chromosome 8 is included in the analysis (p = 0.052, two-tailed Fisher's exact test using pooled data from all four chromosomes).
Figure 6. ICD and Interchromosomal Network (ICN) Models of Chromatin Organization in Mammalian Nuclei
(A) In the ICD model, chromatin from different chromosomes is separated by an ICD compartment rich in nuclear machinery. Active genes are in direct contact with the ICD compartment, as they lie at the surface of CTs or intrachromosomal channels. Rare chromatin loops extending from CTs may invade the ICD space, which is predicted to contain little or no chromatin. Misrejoining between open ends from DSBs on different chromosomes is less likely as broken ends must travel significant distances. (B) In the ICN model, chromatin from different chromosomes is not separated by a compartment but is allowed to expand into the surrounding territories; the presence of adjacent chromosomes, the nuclear membrane, and larger nuclear compartments restricts the amount of intermingling. DNA sequences along chromosomes will have different properties that determine their compaction, mobility, and affinity to specific nuclear components (1). Despite the different local levels of compaction, the global average properties of chromatin in areas of intermingling will be similar to those found within a CT. Rare chromatin loops extending from a CT can invade neighboring CTs (6). Functional associations that correlate with active and inactive states of transcription (2 and 4), including those involving clustering of active RNA polymerases on transcription factories, determine local arrangements within and between chromosomes, that influence the large-scale organization of chromosomes in each cell type. In the ICN model, DSBs formed in regions of intermingling are more likely to produce interchromosomal rearrangements, whereas DSBs elsewhere in the chromosome are more likely to produce intrachromosomal rearrangements.
Comment in
- Interphase chromosomes mingle with their peers.
Chanut F. Chanut F. PLoS Biol. 2006 May;4(5):e174. doi: 10.1371/journal.pbio.0040174. Epub 2006 Apr 25. PLoS Biol. 2006. PMID: 20076580 Free PMC article. No abstract available.
Similar articles
- Chromosome kissing.
Cavalli G. Cavalli G. Curr Opin Genet Dev. 2007 Oct;17(5):443-50. doi: 10.1016/j.gde.2007.08.013. Epub 2007 Oct 22. Curr Opin Genet Dev. 2007. PMID: 17933509 Review. - Spatial relationship between transcription sites and chromosome territories.
Verschure PJ, van Der Kraan I, Manders EM, van Driel R. Verschure PJ, et al. J Cell Biol. 1999 Oct 4;147(1):13-24. doi: 10.1083/jcb.147.1.13. J Cell Biol. 1999. PMID: 10508851 Free PMC article. - Investigation of Spatial Organization of Chromosome Territories in Chromosome Exchange Aberrations After Ionizing Radiation Exposure.
Balajee AS, Sanders JT, Golloshi R, Shuryak I, McCord RP, Dainiak N. Balajee AS, et al. Health Phys. 2018 Jul;115(1):77-89. doi: 10.1097/HP.0000000000000840. Health Phys. 2018. PMID: 29787433 - Mini review: form and function in the human interphase chromosome.
Chevret E, Volpi EV, Sheer D. Chevret E, et al. Cytogenet Cell Genet. 2000;90(1-2):13-21. doi: 10.1159/000015654. Cytogenet Cell Genet. 2000. PMID: 11060439 Review. - Intermingling of chromosome territories.
Szczepińska T, Rusek AM, Plewczynski D. Szczepińska T, et al. Genes Chromosomes Cancer. 2019 Jul;58(7):500-506. doi: 10.1002/gcc.22736. Epub 2019 Mar 3. Genes Chromosomes Cancer. 2019. PMID: 30828902 Review.
Cited by
- Nuclear bodies: the emerging biophysics of nucleoplasmic phases.
Zhu L, Brangwynne CP. Zhu L, et al. Curr Opin Cell Biol. 2015 Jun;34:23-30. doi: 10.1016/j.ceb.2015.04.003. Epub 2015 May 15. Curr Opin Cell Biol. 2015. PMID: 25942753 Free PMC article. Review. - Understanding 3D genome organization by multidisciplinary methods.
Jerkovic I, Cavalli G. Jerkovic I, et al. Nat Rev Mol Cell Biol. 2021 Aug;22(8):511-528. doi: 10.1038/s41580-021-00362-w. Epub 2021 May 5. Nat Rev Mol Cell Biol. 2021. PMID: 33953379 Review. - Chromosome territories have a highly nonspherical morphology and nonrandom positioning.
Khalil A, Grant JL, Caddle LB, Atzema E, Mills KD, Arneodo A. Khalil A, et al. Chromosome Res. 2007;15(7):899-916. doi: 10.1007/s10577-007-1172-8. Epub 2007 Oct 16. Chromosome Res. 2007. PMID: 17926137 - Getting connected in the globin interactome.
Ragoczy T, Groudine M. Ragoczy T, et al. Nat Genet. 2010 Jan;42(1):16-7. doi: 10.1038/ng0110-16. Nat Genet. 2010. PMID: 20037614 - Condensin action and compaction.
Paul MR, Hochwagen A, Ercan S. Paul MR, et al. Curr Genet. 2019 Apr;65(2):407-415. doi: 10.1007/s00294-018-0899-4. Epub 2018 Oct 25. Curr Genet. 2019. PMID: 30361853 Free PMC article. Review.
References
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. - PubMed
- Cremer M, von Hase J, Volm T, Brero A, Kreth G, et al. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–567. - PubMed
- Tanabe H, Habermann FA, Solovei I, Cremer M, Cremer T. Non-random radial arrangements of interphase chromosome territories: Evolutionary considerations and functional implications. Mutat Res. 2002;504:37–45. - PubMed
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, et al. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. - PubMed
- Bridger JM, Boyle S, Kill IR, Bickmore WA. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr Biol. 2000;10:149–152. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources