Polyphyly and gene flow between non-sibling Heliconius species - PubMed (original) (raw)
Polyphyly and gene flow between non-sibling Heliconius species
Vanessa Bull et al. BMC Biol. 2006.
Abstract
Background: The view that gene flow between related animal species is rare and evolutionarily unimportant largely antedates sensitive molecular techniques. Here we use DNA sequencing to investigate a pair of morphologically and ecologically divergent, non-sibling butterfly species, Heliconius cydno and H. melpomene (Lepidoptera: Nymphalidae), whose distributions overlap in Central and Northwestern South America.
Results: In these taxa, we sequenced 30-45 haplotypes per locus of a mitochondrial region containing the genes for cytochrome oxidase subunits I and II (CoI/CoII), and intron-spanning fragments of three unlinked nuclear loci: triose-phosphate isomerase (Tpi), mannose-6-phosphate isomerase (Mpi) and cubitus interruptus (Ci) genes. A fifth gene, dopa decarboxylase (Ddc) produced sequence data likely to be from different duplicate loci in some of the taxa, and so was excluded. Mitochondrial and Tpi genealogies are consistent with reciprocal monophyly, whereas sympatric populations of the species in Panama share identical or similar Mpi and Ci haplotypes, giving rise to genealogical polyphyly at the species level despite evidence for rapid sequence divergence at these genes between geographic races of H. melpomene.
Conclusion: Recent transfer of Mpi haplotypes between species is strongly supported, but there is no evidence for introgression at the other three loci. Our results demonstrate that the boundaries between animal species can remain selectively porous to gene flow long after speciation, and that introgression, even between non-sibling species, can be an important factor in animal evolution. Interspecific gene flow is demonstrated here for the first time in Heliconius and may provide a route for the transfer of switch-gene adaptations for Müllerian mimicry. The results also forcefully demonstrate how reliance on a single locus may give an erroneous picture of the overall genealogical history of speciation and gene flow.
Figures
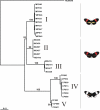
Figure 1
Maximum likelihood genealogy for CoI and CoII. Parsimony bootstrap values (>70%) are given above the nodes, taken from the equivalent nodes on the parsimony trees, when available. Major groups of haplotypes, mostly supported by high bootstrap values or indels are identified using Roman numerals. MG = H. melpomene melpomene (French Guiana), MP = H. melpomene rosina (Panama), CP = H. cydno chioneus (Panama), NUM = H. numata.
Figure 2
Maximum likelihood genealogy for Tpi. Parsimony bootstrap values (> 70%) are given above the nodes, taken from the equivalent nodes on the parsimony trees, when available. Insertions or deletions (indels) inferred to be apomorphies are shown using black bars. Indels inferred to be homoplasious or to involve reversals not concordant with the given topologies, are shown as triangles. Major groups of haplotypes, mostly supported by high bootstrap values or indels are identified using Roman numerals. MG = H. melpomene melpomene (French Guiana), MP = H. melpomene rosina (Panama), CP = H. cydno chioneus (Panama), NUM = H. numata.
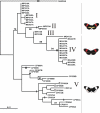
Figure 3
Maximum likelihood genealogy for Mpi. Parsimony bootstrap values (>70%) are given above the nodes, taken from the equivalent nodes on the parsimony trees, when available. Insertions or deletions (indels) inferred to be apomorphies are shown using black bars. Indels inferred to be homoplasious or to involve reversals not concordant with the given topologies, are shown as triangles. Major groups of haplotypes, mostly supported by high bootstrap values or indels are identified using Roman numeral. MG = H. melpomene melpomene (French Guiana), MP = H. melpomene rosina (Panama), CP = H. cydno chioneus (Panama), NUM = H. numata.
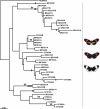
Figure 4
Maximum likelihood genealogy for Ci. Parsimony bootstrap values (>70%) are given above the nodes, taken from the equivalent nodes on the parsimony trees, when available. Insertions or deletions (indels) inferred to be apomorphies are shown using black bars, but indels inferred to be homoplasious or to involve reversals not concordant with the given topologies, are not shown since approximately 27 homoplasious indels would have required showing over 90 gains and losses on the genealogy). This estimated genealogy is poorly resolved and shows many homoplasies, almost certainly due to abundant recombination, and therefore sequence groups were not labeled. MG = H. melpomene melpomene (French Guiana), MP = H. melpomene rosina (Panama), CP = H. cydno chioneus (Panama), NUM = H. numata.
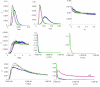
Figure 5
IM analysis of Panama H. melpomene and H. cydno data. The panels show approximate Bayesian posterior probability distributions for effective population size of H. melpomene (θ mel), H. cydno (θ cyd), and the ancestor of the two species (θ A). The time since divergence of the two species (t), and the locus-specific bidirectional introgression rates are also shown (m). The three datasets analysed are the basic IM dataset (blue), and IM reduced dataset 1 (pink) and IM reduced dataset 2 (green). Analysis of the basic IM dataset is compromised by recombination within Tpi and Ci, which is assumed not to occur in the IM algorithm. Reduced datasets containing apparently non-recombined segments of the genes were analyzed to overcome this difficulty. IM reduced dataset 1 differs only from IM reduced dataset 2 in that a different, shorter, part of the Ci locus is used; the low sequence information probably explains why there is little information in the former run in the last panel (probabilities for IM dataset 1 are enhanced 10-fold in this panel only, for clarity). The curves show useful parameter estimation , except in the case of ancestral population size (θ A), the upper tail of the time of divergence (t) and the introgression for Ci for IM reduced dataset 1.
Similar articles
- Phylogenetic discordance at the species boundary: comparative gene genealogies among rapidly radiating Heliconius butterflies.
Beltrán M, Jiggins CD, Bull V, Linares M, Mallet J, McMillan WO, Bermingham E. Beltrán M, et al. Mol Biol Evol. 2002 Dec;19(12):2176-90. doi: 10.1093/oxfordjournals.molbev.a004042. Mol Biol Evol. 2002. PMID: 12446809 - Hybrid speciation in Heliconius butterflies? A review and critique of the evidence.
Brower AV. Brower AV. Genetica. 2011 May;139(5):589-609. doi: 10.1007/s10709-010-9530-4. Epub 2010 Nov 28. Genetica. 2011. PMID: 21113790 Free PMC article. Review. - Historical demography of Mullerian mimicry in the neotropical Heliconius butterflies.
Flanagan NS, Tobler A, Davison A, Pybus OG, Kapan DD, Planas S, Linares M, Heckel D, McMillan WO. Flanagan NS, et al. Proc Natl Acad Sci U S A. 2004 Jun 29;101(26):9704-9. doi: 10.1073/pnas.0306243101. Epub 2004 Jun 21. Proc Natl Acad Sci U S A. 2004. PMID: 15210977 Free PMC article. - Genome-wide patterns of divergence and gene flow across a butterfly radiation.
Nadeau NJ, Martin SH, Kozak KM, Salazar C, Dasmahapatra KK, Davey JW, Baxter SW, Blaxter ML, Mallet J, Jiggins CD. Nadeau NJ, et al. Mol Ecol. 2013 Feb;22(3):814-26. doi: 10.1111/j.1365-294X.2012.05730.x. Epub 2012 Aug 25. Mol Ecol. 2013. PMID: 22924870 - Introgression of wing pattern alleles and speciation via homoploid hybridization in Heliconius butterflies: a review of evidence from the genome.
Brower AV. Brower AV. Proc Biol Sci. 2012 Dec 12;280(1752):20122302. doi: 10.1098/rspb.2012.2302. Print 2013 Feb 7. Proc Biol Sci. 2012. PMID: 23235702 Free PMC article. Review.
Cited by
- Genetic analysis of a wild-caught hybrid between non-sister Heliconius butterfly species.
Dasmahapatra KK, Silva-Vásquez A, Chung JW, Mallet J. Dasmahapatra KK, et al. Biol Lett. 2007 Dec 22;3(6):660-3. doi: 10.1098/rsbl.2007.0401. Biol Lett. 2007. PMID: 17804337 Free PMC article. - How robust are "isolation with migration" analyses to violations of the im model? A simulation study.
Strasburg JL, Rieseberg LH. Strasburg JL, et al. Mol Biol Evol. 2010 Feb;27(2):297-310. doi: 10.1093/molbev/msp233. Epub 2009 Sep 30. Mol Biol Evol. 2010. PMID: 19793831 Free PMC article. - Review. Hybrid trait speciation and Heliconius butterflies.
Jiggins CD, Salazar C, Linares M, Mavarez J. Jiggins CD, et al. Philos Trans R Soc Lond B Biol Sci. 2008 Sep 27;363(1506):3047-54. doi: 10.1098/rstb.2008.0065. Philos Trans R Soc Lond B Biol Sci. 2008. PMID: 18579480 Free PMC article. Review. - Transitions from Single- to Multi-Locus Processes during Speciation with Gene Flow.
Schilling MP, Mullen SP, Kronforst M, Safran RJ, Nosil P, Feder JL, Gompert Z, Flaxman SM. Schilling MP, et al. Genes (Basel). 2018 May 24;9(6):274. doi: 10.3390/genes9060274. Genes (Basel). 2018. PMID: 29795050 Free PMC article. - Selection and demographic history shape the molecular evolution of the gamete compatibility protein bindin in Pisaster sea stars.
Popovic I, Marko PB, Wares JP, Hart MW. Popovic I, et al. Ecol Evol. 2014 May;4(9):1567-88. doi: 10.1002/ece3.1042. Epub 2014 Mar 31. Ecol Evol. 2014. PMID: 24967076 Free PMC article.
References
- Arnold ML. Natural Hybridization and Evolution. Oxford, Oxford University Press; 1997.
- Rieseberg LH. Hybrid origins of plant species. Ann Rev Ecol Syst. 1997;28:359–389. doi: 10.1146/annurev.ecolsys.28.1.359. - DOI
- Dowling TE, Secor CL. The role of hybridization and introgression in the diversification of animals. Ann Rev Ecol Syst. 1997;28:593–620. doi: 10.1146/annurev.ecolsys.28.1.593. - DOI
- Coyne JA, Orr HA. Speciation. Sunderland, Mass., Sinauer Associates; 2004.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous