A kinase-deficient TrkC receptor isoform activates Arf6-Rac1 signaling through the scaffold protein tamalin - PubMed (original) (raw)
A kinase-deficient TrkC receptor isoform activates Arf6-Rac1 signaling through the scaffold protein tamalin
Pedro F Esteban et al. J Cell Biol. 2006.
Abstract
Neurotrophins play an essential role in mammalian development. Most of their functions have been attributed to activation of the kinase-active Trk receptors and the p75 neurotrophin receptor. Truncated Trk receptor isoforms lacking the kinase domain are abundantly expressed during development and in the adult; however, their function and signaling capacity is largely unknown. We show that the neurotrophin-3 (NT3) TrkCT1-truncated receptor binds to the scaffold protein tamalin in a ligand-dependent manner. Moreover, NT3 initiation of this complex leads to activation of the Rac1 GTPase through adenosine diphosphate-ribosylation factor 6 (Arf6). At the cellular level, NT3 binding to TrkCT1-tamalin induces Arf6 translocation to the membrane, which in turn causes membrane ruffling and the formation of cellular protrusions. Thus, our data identify a new signaling pathway elicited by the kinase-deficient TrkCT1 receptor. Moreover, we establish NT3 as an upstream regulator of Arf6.
Figures
Figure 1.
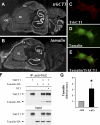
The tamalin PDZ domain interacts with exon 14b of TrkCT1. (A) Schematic representation of the full-length TrkC kinase (TrkC-Kin) and truncated TrkCT1 receptors. EC, extracellular domain; TM, transmembrane domain; JM, juxtamembrane domain. (B) Values of β-galactosidase activity obtained by combining different TrkCT1 bait and tamalin prey plasmids in liquid assay. Note the high β-galactosidase activity when both TrkCT1 exon 14b (aa 575–612) and an intact tamalin PDZ domain are used in the assay (box). Tamalin plasmid “none” indicates that an empty prey plasmid was used. (C) In vitro binding of tamalin to TrkCT1 protein fragments, which contain exon 14b. GST fusion proteins containing COOH-terminal portions of the TrkCT1 receptor were analyzed for their ability to interact in vitro with the full-length tamalin-HA protein by GST pull down-assay, as described in Materials and methods. The presence of tamalin was detected by immunoblotting with an anti-HA antibody (top). Input (bottom) represents 30% of the amount of GST–TrkCT1 fusion proteins (arrows) used in the assay and visualized by Coomassie staining.
Figure 2.
TrkCT1 and tamalin physically interact and colocalize both anatomically and subcellularly. (A and B) In situ hybridization analysis of sagital sections from an adult mouse brain with _trkCT1_- (A) and _tamalin_-specific (B) antisense riboprobes. Note the overlapping pattern of expression in most areas of the forebrain, with particularly high levels in the hippocampus. Hi, hippocampus; Cx, cortex; St, striatum; OB, olfactory bulb. (C–E) HEK293 cells transfected with TrkCT1 and tamalin-HA were stained with the rabbit anti-TrkC antiserum 656 and a mouse monoclonal anti-HA antibody. The secondary antibodies were anti–rabbit rhodamine-conjugated (C; TrkCT1) or anti–mouse fluorescein-conjugated antibodies (D; HA). Fluorescence imaging demonstrates the colocalization of both proteins (E) at the membrane of the cells, as indicated by the yellow color that results from the overlay of red and green fluorescence. (F) Tamalin and TrkCT1 coIP in mammalian cells. HEK293 cells transfected with the indicated plasmid and treated with NT3 were lysed, immunoprecipitated with a goat anti-TrkC antibody, and immunoblotted with anti-HA and anti-TrkC antibody (TrkC 656) as indicated. (top) Western blot of a representative experiment is shown. (bottom) The amount (input) of TrkCT1 and tamalin in 10% of the total cell lysates used for the coIP experiments. (G) Quantification of immunoprecipitated tamalin in double-transfected cells from three independent transfections, each immunoprecipitated and immunoblotted three times (fold difference relative to untreated cells). Error bars represent the mean ± SEM, relative to untreated cells. *, P < 0.05.
Figure 3.
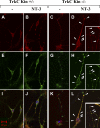
Tamalin and TrkCT1 colocalize in hippocampal neurons in response to NT3. Differentiated control (A, B, E, F, I, and J) and TrkC kinase-deficient (C, D, G, H, K, and L) mouse hippocampal neurons were stained with anti-tamalin– (A–D) and anti-TrkC–specific (E–H) antibodies and subjected to confocal microscopy to investigate tamalin (A–D) and TrkC cellular distribution in the presence (B, F, J, H, D, and L) or absence (A, E, I, C, G, and K) of NT3. Analysis of merged rhodamine and FITC signals reveals colocalization of tamalin and TrkC in response to NT3 treatment (I–L). TrkCT1 has a specific punctuated pattern of expression (G and H; Menn et al., 2000) compared with a more diffuse distribution of the kinase-specific TrkC receptor (E and F). Note the particularly strong punctuated overlapping distribution of tamalin with TrkC in the mutant neurons (D–L, insets) that have only the truncated TrkCT1 receptor (arrowheads).
Figure 4.
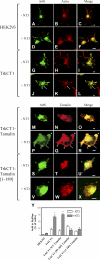
NT3 binding to TrkCT1 induces Arf6 translocation to the cell membrane and ruffling. (A–X) HEK293 (A–F) and HEK293 cells stably expressing TrkCT1 (G–X) were transfected with plasmids directing expression of FLAG-Arf6 (A–X), together with plasmids directing expression of wild-type (M–R) or inactive tamalin (S–X; aa 1–189), and treated with NT3 as indicated (D–F, J–L, P–R, and V–X). Cells were fixed, permeabilized, and stained for Arf6 (green; A, D, G, J, M, P, and S) and polymerized actin (red; B, E, H, and K) or tamalin (red; N, Q, T, and W). Arrows indicate colocalization of Arf6 and actin (J–L), and Arf6 and tamalin (P–R) at the cell membrane in response to NT3 treatment. Arrowheads in P–R indicate cell protrusions caused by NT3 when TrkCT1 and tamalin are coexpressed. (Y) Translocation of Arf6 to the membrane of TrkCT1-expressing cells in response to NT3 is inhibited by dominant-negative forms of tamalin. Percentage of cells with Arf6 at the cell edge in either protrusions or ruffles based on counts of at least 50 cells in each of two separate experiments. Two separate dominant-negative tamalin (aa 99–189 and aa 1–189) were used. Error bars represent the mean ± SEM, relative to untreated cells. n ≥ 100. **, P < 0.01; ***, P < 0.001. ANOVA followed by a Tukey test were used for both. Bars, 5 μm.
Figure 5.
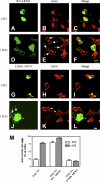
Cytohesin-2–ARNO mediates NT3-induced membrane ruffling and actin protrusion formation. HEK293 cells expressing TrkCT1 were transfected with a wild-type HA-tagged (WT-ARNO; A–F) or a catalytically inactive Myc-tagged ARNO (E156K-ARNO; G–L) and stained for polymerized actin. Ruffling, as indicated by the extensive presence of polymerized actin, is seen in cells expressing WT-ARNO in response to NT3 (D–F and arrows in E). On the contrary, the catalytically inactive E156-ARNO disrupts actin polymerization in response to NT3 (K), although it gets recruited to the cell edge because it can still bind tamalin (J, arrows). Arrowheads in K indicate ruffling in cells that do not express the inactive E156-ARNO, but are still able to use the endogenous ARNO for NT3 signaling. (M) Percentage of cells with actin at the cell edge in response to NT3 was determined from untransfected (TrkC.T1) or transfected (TrkC.T1-WT ARNO and TrkC.T1–E156K-ARNO) cells from two separate experiments. Error bars represent the mean ± SEM. Bars, 10 μm.
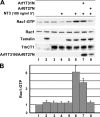
Figure 6.
NT3 induces Rac1 activation via the TrkCT1–tamalin–Arf6 pathway. (A) HEK293 cells expressing TrkCT1 (lane 3), tamalin-HA (lane 2), and TrkCT1 and tamalin-HA (lanes 5–8) in the absence (lanes 5 and 6) or presence of inactive Arf1 (lane 7) or Arf6 (lane 8) were serum starved for 13 h and stimulated with 100 ng/ml of NT3 for 5 min, where indicated. Relative Rac1 activity to total Rac1 protein normalized to the basal activity present in untransfected HEK293 cells is shown. Input for Rac1, tamalin, TrkCT1, and Arf1T31N or Arf6T27N represents 1% of the total cell extract used to detect the active Rac1-GTP (top). (B) Quantification of Rac1 activation in response to NT3 treatment. The fold difference is mean ± SEM, relative to untreated cells (lane 1). Error bars represent the mean ± SEM from six independent experiments, except for the experiments performed with the Arf1 or Arf6 dominant-negative forms (three experiments).
Figure 7.
Schematic representation of the newly identified NT3-activated signaling pathway. NT3 binding to TrkCT1 causes the recruitment of tamalin to the TrkCT1 cytoplasmic domain (exon 14b). In turn, tamalin activates the guanine nucleotide exchange factors cytohesin-2–ARNO (Kitano et al., 2002), which causes the switch of Arf6 from its inactive to its active Arf6-GTP form. Arf6-GTP induces Rac1 activation and is translocated to the membrane, where ruffling and actin reorganization take place. Tamalin alanine/proline (A/P), PDZ, leucine rich (Leu), and PDZ-binding (SQL) domains are indicated, as well as ARNO's pleckstrin homology (PH), coiled coil, and Sec7 homology domains.
Similar articles
- ARF1 and ARF6 regulate recycling of GRASP/Tamalin and the Rac1-GEF Dock180 during HGF-induced Rac1 activation.
Koubek EJ, Santy LC. Koubek EJ, et al. Small GTPases. 2018 May 4;9(3):242-259. doi: 10.1080/21541248.2016.1219186. Epub 2016 Aug 26. Small GTPases. 2018. PMID: 27562622 Free PMC article. - ARAP2 signals through Arf6 and Rac1 to control focal adhesion morphology.
Chen PW, Jian X, Yoon HY, Randazzo PA. Chen PW, et al. J Biol Chem. 2013 Feb 22;288(8):5849-60. doi: 10.1074/jbc.M112.415778. Epub 2013 Jan 7. J Biol Chem. 2013. PMID: 23295182 Free PMC article. - In vitro and in vivo analysis of neurotrophin-3 activation of Arf6 and Rac-1.
Esteban PF, Caprari P, Yoon HY, Randazzo PA, Tessarollo L. Esteban PF, et al. Methods Enzymol. 2008;438:171-83. doi: 10.1016/S0076-6879(07)38012-9. Methods Enzymol. 2008. PMID: 18413248 Free PMC article. - ARF6-Rac1 signaling-mediated neurite outgrowth is potentiated by the neuronal adaptor FE65 through orchestrating ARF6 and ELMO1.
Chan WWR, Li W, Chang RCC, Lau KF. Chan WWR, et al. FASEB J. 2020 Dec;34(12):16397-16413. doi: 10.1096/fj.202001703R. Epub 2020 Oct 13. FASEB J. 2020. PMID: 33047393 - ARF6 is required for growth factor- and rac-mediated membrane ruffling in macrophages at a stage distal to rac membrane targeting.
Zhang Q, Calafat J, Janssen H, Greenberg S. Zhang Q, et al. Mol Cell Biol. 1999 Dec;19(12):8158-68. doi: 10.1128/MCB.19.12.8158. Mol Cell Biol. 1999. PMID: 10567541 Free PMC article.
Cited by
- GRASP and IPCEF promote ARF-to-Rac signaling and cell migration by coordinating the association of ARNO/cytohesin 2 with Dock180.
White DT, McShea KM, Attar MA, Santy LC. White DT, et al. Mol Biol Cell. 2010 Feb 15;21(4):562-71. doi: 10.1091/mbc.e09-03-0217. Epub 2009 Dec 16. Mol Biol Cell. 2010. PMID: 20016009 Free PMC article. - Postsynaptic TrkC and presynaptic PTPσ function as a bidirectional excitatory synaptic organizing complex.
Takahashi H, Arstikaitis P, Prasad T, Bartlett TE, Wang YT, Murphy TH, Craig AM. Takahashi H, et al. Neuron. 2011 Jan 27;69(2):287-303. doi: 10.1016/j.neuron.2010.12.024. Neuron. 2011. PMID: 21262467 Free PMC article. - Integrative epigenomic and genomic filtering for methylation markers in hepatocellular carcinomas.
Shen J, LeFave C, Sirosh I, Siegel AB, Tycko B, Santella RM. Shen J, et al. BMC Med Genomics. 2015 Jun 10;8:28. doi: 10.1186/s12920-015-0105-1. BMC Med Genomics. 2015. PMID: 26059414 Free PMC article. - Physiological and Pathological Roles of the Cytohesin Family in Neurons.
Ito A, Fukaya M, Okamoto H, Sakagami H. Ito A, et al. Int J Mol Sci. 2022 May 3;23(9):5087. doi: 10.3390/ijms23095087. Int J Mol Sci. 2022. PMID: 35563476 Free PMC article. Review. - Extracellular regulation of type IIa receptor protein tyrosine phosphatases: mechanistic insights from structural analyses.
Coles CH, Jones EY, Aricescu AR. Coles CH, et al. Semin Cell Dev Biol. 2015 Jan;37:98-107. doi: 10.1016/j.semcdb.2014.09.007. Epub 2014 Sep 16. Semin Cell Dev Biol. 2015. PMID: 25234613 Free PMC article. Review.
References
- Aramori, I., and S. Nakanishi. 1992. Signal transduction and pharmacological characteristics of a metabotropic glutamate receptor, mGluR1, in transfected CHO cells. Neuron. 8:757–765. - PubMed
- Cohen-Cory, S., and S.E. Fraser. 1995. Effects of brain-derived neurotrophic factor on optic axon branching and remodelling in vivo. Nature. 378:192–196. - PubMed
- Donaldson, J.G. 2003. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J. Biol. Chem. 278:41573–41576. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials