Highly conserved syntenic blocks at the vertebrate Hox loci and conserved regulatory elements within and outside Hox gene clusters - PubMed (original) (raw)
Highly conserved syntenic blocks at the vertebrate Hox loci and conserved regulatory elements within and outside Hox gene clusters
Alison P Lee et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Hox genes in vertebrates are clustered, and the organization of the clusters has been highly conserved during evolution. The conservation of Hox clusters has been attributed to enhancers located within and outside the Hox clusters that are essential for the coordinated "temporal" and "spatial" expression patterns of Hox genes in developing embryos. To identify evolutionarily conserved regulatory elements within and outside the Hox clusters, we obtained contiguous sequences for the conserved syntenic blocks from the seven Hox loci in fugu and carried out a systematic search for conserved noncoding sequences (CNS) in the human, mouse, and fugu Hox loci. Our analysis has uncovered unusually large conserved syntenic blocks at the HoxA and HoxD loci. The conserved syntenic blocks at the human and mouse HoxA and HoxD loci span 5.4 Mb and 4 Mb and contain 21 and 19 genes, respectively. The corresponding regions in fugu are 16- and 12-fold smaller. A large number of CNS was identified within the Hox clusters and outside the Hox clusters spread over large regions. The CNS include previously characterized enhancers and overlap with the 5' global control regions of HoxA and HoxD clusters. Most of the CNS are likely to be control regions involved in the regulation of Hox and other genes in these loci. We propose that the regulatory elements spread across large regions on either side of Hox clusters are a major evolutionary constraint that has maintained the exceptionally long syntenic blocks at the HoxA and HoxD loci.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Fig. 1.
Conserved syntenic blocks at the fugu and human Hox loci. Fugu has two duplicate loci for human HoxA, HoxB, and HoxD loci. Hexagons represent genes. MicroRNA genes are shown as borderless hexagons and pseudogenes with dotted borders. Genes that are present in only fugu or human locus are shown as open hexagons. Dark colored hexagons in the fugu (linked with their human orthologs with a diagonal line) represent genes that have undergone rearrangements.
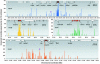
Fig. 2.
Profiles of CNS at the human HoxA, HoxB, HoxC, and HoxD loci. x axis represents chromosomal coordinates, and y axis represents CNS. Genes at each locus are shown at the top as red boxes (Hox genes) or open boxes (non-Hox genes) linked with a thin line. Names of some genes are indicated. The previously identified 5′ global enhancers at HoxD and HoxA loci (3, 5) are represented by purple oval shapes.
Similar articles
- Elephant shark (Callorhinchus milii) provides insights into the evolution of Hox gene clusters in gnathostomes.
Ravi V, Lam K, Tay BH, Tay A, Brenner S, Venkatesh B. Ravi V, et al. Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16327-32. doi: 10.1073/pnas.0907914106. Epub 2009 Sep 3. Proc Natl Acad Sci U S A. 2009. PMID: 19805301 Free PMC article. - Evolutionary conservation of regulatory elements in vertebrate Hox gene clusters.
Santini S, Boore JL, Meyer A. Santini S, et al. Genome Res. 2003 Jun;13(6A):1111-22. doi: 10.1101/gr.700503. Genome Res. 2003. PMID: 12799348 Free PMC article. - Hoxb-13: a new Hox gene in a distant region of the HOXB cluster maintains colinearity.
Zeltser L, Desplan C, Heintz N. Zeltser L, et al. Development. 1996 Aug;122(8):2475-84. doi: 10.1242/dev.122.8.2475. Development. 1996. PMID: 8756292 - Ancestral and recently recruited global control of the Hox genes in development.
Deschamps J. Deschamps J. Curr Opin Genet Dev. 2007 Oct;17(5):422-7. doi: 10.1016/j.gde.2007.07.008. Epub 2007 Sep 17. Curr Opin Genet Dev. 2007. PMID: 17870464 Review. - Why are Hox genes clustered?
Mann RS. Mann RS. Bioessays. 1997 Aug;19(8):661-4. doi: 10.1002/bies.950190804. Bioessays. 1997. PMID: 9264246 Review.
Cited by
- Evolutionary divergence of a Hoxa2b hindbrain enhancer in syngnathids mimics results of functional assays.
Fuiten AM, Cresko WA. Fuiten AM, et al. Dev Genes Evol. 2021 Jul;231(3-4):57-71. doi: 10.1007/s00427-021-00676-x. Epub 2021 May 18. Dev Genes Evol. 2021. PMID: 34003345 - Comparative genomic analysis of prion genes.
Premzl M, Gamulin V. Premzl M, et al. BMC Genomics. 2007 Jan 2;8:1. doi: 10.1186/1471-2164-8-1. BMC Genomics. 2007. PMID: 17199895 Free PMC article. - Are we degenerate tetraploids? More genomes, new facts.
Abbasi AA. Abbasi AA. Biol Direct. 2008 Dec 10;3:50. doi: 10.1186/1745-6150-3-50. Biol Direct. 2008. PMID: 19077184 Free PMC article. - Sequence determinants, function, and evolution of CpG islands.
Angeloni A, Bogdanovic O. Angeloni A, et al. Biochem Soc Trans. 2021 Jun 30;49(3):1109-1119. doi: 10.1042/BST20200695. Biochem Soc Trans. 2021. PMID: 34156435 Free PMC article. Review. - HoxD expression in the fin-fold compartment of basal gnathostomes and implications for paired appendage evolution.
Tulenko FJ, Augustus GJ, Massey JL, Sims SE, Mazan S, Davis MC. Tulenko FJ, et al. Sci Rep. 2016 Mar 4;6:22720. doi: 10.1038/srep22720. Sci Rep. 2016. PMID: 26940624 Free PMC article.
References
- Kmita M., Duboule D. Science. 2003;301:331–333. - PubMed
- Chambeyron S., Da Silva N. R., Lawson K. A., Bickmore W. A. Development (Cambridge, U.K.) 2005;132:2215–2223. - PubMed
- Spitz F., Gonzalez F., Duboule D. Cell. 2003;113:405–417. - PubMed
- Zakany J., Kmita M., Duboule D. Science. 2004;304:1669–1672. - PubMed
- Lehoczky J. A., Williams M. E., Innis J. W. Evol. Dev. 2004;6:423–430. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous