Amino acids regulate retrieval of the yeast general amino acid permease from the vacuolar targeting pathway - PubMed (original) (raw)
Amino acids regulate retrieval of the yeast general amino acid permease from the vacuolar targeting pathway
Marta Rubio-Texeira et al. Mol Biol Cell. 2006 Jul.
Abstract
Intracellular sorting of the general amino acid permease (Gap1p) in Saccharomyces cerevisiae depends on availability of amino acids such that at low amino acid concentrations Gap1p is sorted to the plasma membrane, whereas at high concentrations Gap1p is sorted to the vacuole. In a genome-wide screen for mutations that affect Gap1p sorting we identified deletions in a subset of components of the ESCRT (endosomal sorting complex required for transport) complex, which is required for formation of the multivesicular endosome (MVE). Gap1p-GFP is delivered to the vacuolar interior by the MVE pathway in wild-type cells, but when formation of the MVE is blocked by mutation, Gap1p-GFP efficiently cycles from this compartment to the plasma membrane, resulting in unusually high permease activity at the cell surface. Importantly, cycling of Gap1p-GFP to the plasma membrane is blocked by high amino acid concentrations, defining recycling from the endosome as a major step in Gap1p trafficking under physiological control. Mutations in LST4 and LST7 genes, previously identified for their role in Gap1p sorting, similarly block MVE to plasma membrane trafficking of Gap1p. However, mutations in other recycling complexes such as the retromer had no significant effect on the intracellular sorting of Gap1p, suggesting that Gap1p follows a genetically distinct pathway for recycling. We previously found that Gap1p sorting from the Golgi to the endosome requires ubiquitination of Gap1p by an Rsp5p ubiquitin ligase complex, but amino acid abundance does not appear to significantly alter the accumulation of polyubiquitinated Gap1p. Thus the role of ubiquitination appears to be a signal for delivery of Gap1p to the MVE, whereas amino acid abundance appears to control the cycling of Gap1p from the MVE to the plasma membrane.
Figures
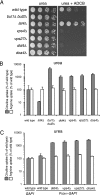
Figure 1.
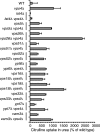
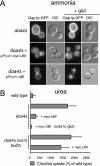
Null mutations in class E vps genes cause increased activity of Gap1p. (A) Wild-type (CKY835), lst4Δ (CKY695), bul1Δ bul2Δ (CKY698), did4Δ (CKY839), vps4Δ (CKY836), vps27Δ (CKY837), and doa4Δ (CKY838) strains were spotted as serial dilutions onto minimal urea medium, with or without 7 mg/l of ADCB. (B) The same strains were grown in liquid minimal urea medium and assayed for [14C]citrulline and [14C]arginine uptake. (C) Wild-type (CKY835) and strains carrying a genomic PADH1-GAP1 replacement of the endogenous GAP1 gene (CKY833), combined with did4Δ (CKY840), vps4Δ (CKY841), vps27Δ (CKY842), and doa4Δ (CKY843) mutations, were assayed for [14C]citrulline and [14C]arginine uptake. The data are expressed as a percentage of the rate of uptake for the wild-type strain and represent the mean for at least four independent experiments. Error bars, 1 SD.
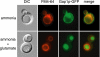
Figure 2.
Gap1p is sorted into the vacuolar lumen. A wild-type strain expressing a genomic GFP-tagged version of GAP1 (CKY834) was grown to exponential phase in minimal ammonia medium. Glutamate was added to a final concentration of 3 mM, and cells were incubated for an additional hour. Cells were then labeled with the vacuolar membrane staining dye FM4-64 for 30 min, followed by a 60-min chase before microscopy. Cells were imaged using a UV fluorescence microscope with a GFP filter (GFP), a rhodamine filter (FM4-64), and by Nomarski optics (DIC).
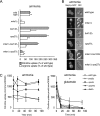
Figure 3.
Inactivation of Gap1p activity by amino acids occurs in the class E vps mutants but not in mutants impaired for Gap1p ubiquitination. (A) [14C]citrulline and [14C]arginine uptake as assayed in a mutant with increased levels of catabolic amino acids, mks1Δ (CKY758), combined with the mutant background defective in Gap1p polyubiquitination, bul1Δ bul2Δ (mks1Δ bul1Δ bul2Δ; CKY923) or the mutant class E background, vps27Δ (mks1Δ vps27Δ; CKY924). Activities for the wild-type strain (CKY835) and the mutant controls bul1Δ bul2Δ (CKY698) and vps27Δ (CKY837) grown in parallel are provided for comparison. The cell cultures were grown in minimal ammonia medium. Averaged data are expressed as in Figure 1. (B) Gap1p-GFP localization is shown in cells from the mutants mks1Δ (CKY867), mks1Δ bul1Δ bul2Δ (CKY926), and mks1Δ vps27Δ (CKY927), grown in minimal medium with ammonia as a nitrogen source. Gap1p-GFP images of the wild-type strain (CKY834), the mutant controls bul1Δ bul2Δ (CKY925), and vps27Δ (CKY851), grown identically, are shown for comparison. (C) A [14C]citrulline uptake time course from the following strains: wild-type (CKY835; □), end3Δ (CKY934; ◇), did4Δ (CKY839; ▴), vps4Δ (CKY836; ■), and vps27Δ (CKY837; ♦), growing in liquid minimal ammonia medium (left panel) or ammonia medium plus 3 mM glutamate, added when the strains were at OD600 of 0.2/ml (right panel), is shown. These graphics show the averaged data between three independent experiments.
Figure 4.
The lst4Δ mutation has a similar effect on Gap1p trafficking as glutamate and high amino acid concentrations. The strains doa4Δ (CKY838) and lst4Δ doa4Δ (CKY847), bro1Δ (CKY935) and lst4Δ bro1Δ (CKY936), did4Δ (CKY839) and lst4Δ did4Δ (CKY844), vps4Δ (CKY836) and lst4Δ vps4Δ (CKY845), vps27Δ (CKY837) and lst4Δ vps27Δ (CKY846), were spotted as serial dilutions onto minimal ammonia medium, with or without a sub-LC of ADCB (7 mg/l). As control strains, a wild-type strain (CKY835) and the mutants lst4Δ (CKY695), bul1Δ bul2Δ (CKY698), and lst4Δ bul1Δ bul2Δ (CKY699) grown in identical conditions, are shown in the top panel. (B) The same strains growing in minimal urea medium were assayed for [14C]citrulline uptake. Averaged data are expressed as in Figure 1.
Figure 5.
Class E VPS mutants accumulate Gap1p-GFP in endosomal and perivacuolar membranes in the presence of glutamate or an lst4Δ mutation. (A) Fluorescence microscopy images of wild-type cells (CKY834) and the mutants lst4Δ (CKY848), vps4Δ (CKY850), vps27Δ (CKY851), did4Δ (CKY849), bro1Δ (CKY937), lst4Δ vps4Δ (CKY854), lst4Δ vps27Δ (CKY855; leucine auxotrophy covered by the CEN-LEU2 plasmid pRS415), lst4Δ did4Δ (CKY853), lst4Δ bro1Δ (CKY938), expressing genomic GFP-tagged Gap1p and exponentially growing in minimal ammonia medium, or incubated for 1 h in the presence of 3 mM glutamate before imaging. (B) A subset of class E vps mutations causes accumulation of Gap1p-GFP in endosomal and perivacuolar membranes in the absence of high concentrations of amino acids. Cells from wild-type strain (CKY835) and the mutants vps27Δ (CKY837), did2Δ (CKY857), hse1Δ (CKY858), vps23Δ (CKY859), vps37Δ (CKY860), and vps60Δ (CKY861), expressing Gap1p-GFP from a centromeric plasmid (p_GAP1-GFP_) and exponentially growing in minimal ammonia medium were imaged by fluorescence microscopy.
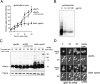
Figure 6.
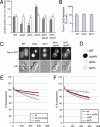
Gap1p accumulated in the prevacuolar compartment of a class E vps mutant can recycle to the plasma membrane. (A) The strains vps4Δ (CKY836) and vps4Δ lst4Δ (CKY845), grown in minimal glutamate medium, were transferred to minimal urea medium and immediately assayed for [14C]citrulline uptake at different times after the shift. Activity time courses are represented as filled diamonds (vps4Δ) or filled triangles (vps4Δ lst4Δ). The strain vps4Δ was also assayed in the presence of 1.5 μg/ml cycloheximide in the urea-containing medium before shift, to inhibit translation of newly synthesized Gap1p (◇). (B) The effect of cycloheximide (0, 1.5, 10, and 100 μg/ml) on bulk translation was assayed by pulse labeling vps4Δ (CKY836) on minimal glutamate medium with [35S]methionine. (C) Protein extracts taken at the same time-point periods and using the same strains and conditions as in A, were subject to SDS-PAGE and Western blotting with Gap1p antibody. As a loading control, Pgk1p levels are shown (bottom blot). Each lane contains an extract from the same number of cells. (D) The strains vps4Δ (CKY850) and lst4Δ vps4Δ (CKY854) were monitored for Gap1p-GFP localization at different periods of time after being shifted from minimal glutamate medium to urea medium. For comparison, images of the same strains steadily growing in urea are also shown.
Figure 7.
Recycling of Gap1p is not severely affected by defects in pathways involved in the recycling of proteins from the MVE. The following strains were grown in liquid minimal urea medium and assayed for [14C]citrulline uptake: wild type (CKY835), lst4Δ (CKY695), vps4Δ (CKY836), lst4Δ vps4Δ (CKY845), vps5Δ (Y01845), vps5Δ vps4Δ (CKY995), vps26Δ (Y01370), vps26Δ vps4Δ (CKY996), vps51Δ (Y05091), vps51Δ vps4Δ (CKY997), vps52Δ (Y04318), vps52Δ vps4Δ (CKY998), vps16Δ (Y02783), vps16Δ vps4Δ (CKY999), vps18Δ (Y04105), vps18Δ vps4Δ (CKY1000), ypt6Δ (Y05171), ypt6Δ vps4Δ (CKY1001), ypt7Δ (Y00575), ypt7Δ vps4Δ (CKY1002), vam3Δ (Y02362), vam3Δ vps4Δ (CKY1003), vps33Δ (Y05305), vps33Δ vps4Δ (CK1004). Measurements of Gap1p activity are expressed as in Figure 1. Single mutant strains in the BY4741 background (EUROSCARF deletion strains), and some of the double mutant strains were transformed with the centromeric plasmid p_CEN_-HIS3-LEU2-MET15 (pCK283) to eliminate auxotrophic requirements for amino acids.
Figure 8.
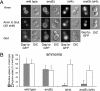
LST4 and LST7 are required specifically for Gap1p sorting. (A) lst4Δ and lst7Δ mutations interfere with Gap1p distribution to the plasma membrane caused by a vps4Δ mutation. The following strains were grown in minimal urea medium and assayed for uptake of [14C]citrulline and [14C]arginine: wild type (CKY835), lst4Δ (CKY695), lst7Δ (CKY994), vps4Δ (CKY836), lst4Δ vps4Δ (CKY845), and lst7Δ vps4Δ (CKY1005). (B) The decrease in Gap1p activity caused by lst4Δ and lst7Δ mutations is not a transcriptional effect. Wild type (CKY835), lst4Δ (CKY695), and lst7Δ (CKY994) strains were transformed with the PGAP1-LacZ (pMS29) plasmid and assayed for β-galactosidase activity after growth in minimal urea medium. (C) lst4Δ and lst7Δ mutations cause constitutive sorting of Gap1p to the vacuole and block its recycling caused by a vps4Δ mutation. Cells from wild-type strain (CKY835) and the mutants lst4Δ (CKY695), lst7Δ (CKY994), lst4Δ vps4Δ (CKY845), and lst7Δ vps4Δ (CKY1005), expressing Gap1p-GFP from a centromeric plasmid (p_GAP1-GFP_) and exponentially growing in minimal ammonia medium were visualized by fluorescence microscopy. (D) Wild-type (CKY835) vps4Δ (CKY836), lst4Δ (CKY695), and lst7Δ (CKY994) were spotted on a nitrocellulose membrane on YPD plates (an equivalent to 0.5 OD600 of cells/spot) and after 16 h at 30°C secreted CPY was detected using monoclonal anti-CPY (Molecular Probes). (E and F) lst4Δ and lst7Δ mutations do not interfere with recycling of FM4-64. (E) The wild-type strain BY4741 was assayed for the ability to recycle the fluorescent membrane dye FM4–64 as explained in Materials and Methods. Fluorescence was measured every second for 10 min on a spectrofluorometer and the graphic represents the average of three independent experiments. As a negative control the same strain was treated with 10 mM NaN3 to block vesicular trafficking, and as a positive control the mutant strain from identical genetic background, rcy1Δ (Y01221) was also assayed. (F) The same experiment as in E was carried out in parallel in the wild-type (CKY835), lst4Δ (CKY695), and lst7Δ (CKY994) strains.
Figure 9.
A lst4Δ mutation or growth on glutamate causes redistribution of Gap1p independently of endocytosis. Fluorescence microscopy images from cells of the wild-type (CKY834) strain, the mutant lst4Δ (CKY848), and the mutants impaired in endocytosis end3Δ (CKY874), and end3Δ lst4Δ (CKY875), expressing genomic GFP-tagged Gap1p, are shown. Cells were continuously grown to exponential phase in minimal medium with ammonia (top panel) or glutamate (bottom panel) as the only nitrogen source. Images of cells taken 30 min after the addition of glutamate 3 mM to induce endocytosis of Gap1p-GFP are also shown (middle panel). (B) The same strains were grown in liquid minimal ammonia medium and assayed for [14C]citrulline and [14C]arginine uptake, and the data were averaged as in Figure 1.
Figure 10.
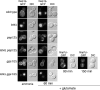
Constitutive vacuolar sorting of Gap1p in a lst4Δ mutant does not depend on GGA function. The following strains were transformed with the centromeric plasmid (p_GAP1-GFP_) and imaged by fluorescence microscopy to detect Gap1p-GFP localization: Wild-type (CKY835), lst4Δ (CKY695), pep12Δ (CKY694), lst4Δ pep12Δ (CKY1006), gga1Δ gga2Δ (CKY1007), and lst4Δ gga1Δ gga2Δ (CKY1008). Cells were continuously grown in ammonia or shifted from ammonia to glutamate.
Figure 11.
Overexpression of ubiquitin restores Gap1p vacuolar sorting in a doa4Δ mutant. (A) Gap1p-GFP localization in cells grown in minimal ammonia medium was monitored by fluorescence microscopy in a doa4Δ mutant (CKY852) alone or transformed with the PCUP1-myc-UBI plasmid. Glutamate was added and cells were imaged after 30 min of incubation. The same experiment carried out by transformation with the PCUP1-UBI plasmid is also shown. (B) Gap1p activity ([14C]citrulline uptake rate) in strains constitutively expressing genomic HA-tagged Gap1p from the constitutive promoter ADH1 was measured from cultures grown in minimal urea medium. The activity of a wild-type (CKY868) strain is compared with that of a doa4Δ mutant (CKY928) alone or transformed with the _PCUP1-myc_-UBI plasmid. The ability of doa4Δ (CKY928) containing PCUP1-myc-UBI compared with the inability of a doa4Δ bul1Δ bul2Δ (CKY930), carrying this same plasmid, to down-regulate Gap1p activity is also shown (shift from minimal urea medium to glutamate medium for 1 h). Averaged data are represented as in Figure 1.
Figure 12.
An lst4Δ mutation, as growth in glutamate, does not cause an increased accumulation in polyubiquitinated Gap1p comparable to the overproduction of Bul1p. (A) The polyubiquitinated state of Gap1p in the wild-type strain (lane 3, CKY868) versus a doa4Δ mutant (lane 4, CKY928) in a pep4Δ background expressing genomic PADH1-GAP1-HA and transformed with p_PCUP1-myc-UBI_, was compared in immunoprecipitates from cells grown in minimal urea medium. Identical backgrounds (lane 1, CKY474, and lane 2, CKY993, respectively) expressing endogenous untagged Gap1p were utilized as negative controls. Immunoprecipitated samples were prepared as described in Materials and Methods, normalized, and 1/20 of the total loaded for SDS-PAGE and immunoblotting. Anti-HA (rat, 3F10) immunoprecipitates were immunoblotted with either anti-HA (16B12) mouse (left panel) or anti-myc (9E10) mouse (right panel). (B) Polyubiquitinated Gap1p-HA was isolated from pep4Δ PADH1-GAP1-HA strains containing the plasmid p_PCUP1-myc_-UBI and 1/40 of the total normalized sample was loaded for SDS-PAGE and immunoblotted as in A. Samples from the strains analyzed are presented in the blots as follows: doa4Δ (lane 2, CKY928), doa4Δ gap1 K9R, K16R -HA (lane 3, CKY929), doa4Δ bul1Δ bul2Δ (lane 4, CKY930), doa4Δ lst4Δ (lane 6, CKY931), and doa4Δ (CKY1009) transformed with the empty plasmid pRS423 (lane 5), or the Bul1p overproducing strain pBUL1 (lane 7), and doa4Δ (CKY928) shifted for 1 h from urea to glutamate as the only nitrogen source (lane 8). A doa4Δ mutant strain expressing endogenous Gap1p served as a negative control (lane 1, CKY993).
Figure 13.
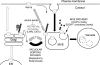
Proposed model for regulated Gap1p sorting in the endosome. Newly synthesized Gap1p has two possible fates once it reaches the _trans_-Golgi: sorting to the plasma membrane where it is active for amino acid uptake or to the vacuole for degradation. Polyubiquitination mediated by the Rsp5/Bul1p/Bul2p ubiquitin ligase complex is necessary for Gap1p delivery to the vacuolar sorting pathway (arrows in black). Polyubiquitinated Gap1p reaches the prevacuolar compartment where it has two possible fates: to become cargo for entry into multivesicular endosomes (MVE) or for recycling to the plasma membrane by a trafficking step that requires Lst4p and Lst7p (gray arrows). Recycling of Gap1p may occur by direct trafficking from the MVE to the plasma membrane or by way of the _trans_-Golgi. High intracellular concentrations of amino acids block Gap1p retrieval from the MVE. Before its delivery into the luminal vesicles of the MVE, Gap1p undergoes Doa4-dependent deubiquitination. By regenerating free ubiquitin, Doa4p also controls the rate of Rsp5/Bul1/Bul2-dependent polyubiquitination of newly synthesized Gap1p, earlier in the pathway. The MVE finally fuses to the vacuole, releasing the Gap1p-containing vesicles into the vacuolar lumen for degradation. Mutants that block formation of MVE vesicles (such as vps4Δ or vps27Δ) cause most Gap1p to follow the recycling pathway to the plasma membrane.
Similar articles
- Transport activity-dependent intracellular sorting of the yeast general amino acid permease.
Cain NE, Kaiser CA. Cain NE, et al. Mol Biol Cell. 2011 Jun 1;22(11):1919-29. doi: 10.1091/mbc.E10-10-0800. Epub 2011 Apr 6. Mol Biol Cell. 2011. PMID: 21471002 Free PMC article. - Different ubiquitin signals act at the Golgi and plasma membrane to direct GAP1 trafficking.
Risinger AL, Kaiser CA. Risinger AL, et al. Mol Biol Cell. 2008 Jul;19(7):2962-72. doi: 10.1091/mbc.e07-06-0627. Epub 2008 Apr 23. Mol Biol Cell. 2008. PMID: 18434603 Free PMC article. - Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by SEC13, LST4, LST7 and LST8.
Roberg KJ, Bickel S, Rowley N, Kaiser CA. Roberg KJ, et al. Genetics. 1997 Dec;147(4):1569-84. doi: 10.1093/genetics/147.4.1569. Genetics. 1997. PMID: 9409822 Free PMC article. - The ubiquitin code of yeast permease trafficking.
Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, André B. Lauwers E, et al. Trends Cell Biol. 2010 Apr;20(4):196-204. doi: 10.1016/j.tcb.2010.01.004. Trends Cell Biol. 2010. PMID: 20138522 Review. - Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae.
Bowers K, Stevens TH. Bowers K, et al. Biochim Biophys Acta. 2005 Jul 10;1744(3):438-54. doi: 10.1016/j.bbamcr.2005.04.004. Biochim Biophys Acta. 2005. PMID: 15913810 Review.
Cited by
- Regulation of Sensing, Transportation, and Catabolism of Nitrogen Sources in Saccharomyces cerevisiae.
Zhang W, Du G, Zhou J, Chen J. Zhang W, et al. Microbiol Mol Biol Rev. 2018 Feb 7;82(1):e00040-17. doi: 10.1128/MMBR.00040-17. Print 2018 Jun. Microbiol Mol Biol Rev. 2018. PMID: 29436478 Free PMC article. Review. - A split-ubiquitin two-hybrid screen for proteins physically interacting with the yeast amino acid transceptor Gap1 and ammonium transceptor Mep2.
Van Zeebroeck G, Kimpe M, Vandormael P, Thevelein JM. Van Zeebroeck G, et al. PLoS One. 2011;6(9):e24275. doi: 10.1371/journal.pone.0024275. Epub 2011 Sep 2. PLoS One. 2011. PMID: 21912684 Free PMC article. - FLCN: The causative gene for Birt-Hogg-Dubé syndrome.
Schmidt LS, Linehan WM. Schmidt LS, et al. Gene. 2018 Jan 15;640:28-42. doi: 10.1016/j.gene.2017.09.044. Epub 2017 Sep 29. Gene. 2018. PMID: 28970150 Free PMC article. Review. - A calcineurin-dependent switch controls the trafficking function of α-arrestin Aly1/Art6.
O'Donnell AF, Huang L, Thorner J, Cyert MS. O'Donnell AF, et al. J Biol Chem. 2013 Aug 16;288(33):24063-80. doi: 10.1074/jbc.M113.478511. Epub 2013 Jul 3. J Biol Chem. 2013. PMID: 23824189 Free PMC article. - Activity-dependent reversible inactivation of the general amino acid permease.
Risinger AL, Cain NE, Chen EJ, Kaiser CA. Risinger AL, et al. Mol Biol Cell. 2006 Oct;17(10):4411-9. doi: 10.1091/mbc.e06-06-0506. Epub 2006 Aug 2. Mol Biol Cell. 2006. PMID: 16885415 Free PMC article.
References
- Adams A., Gottschling D., Kaiser C. Methods in Yeast Genetics: A Laboratory Course Manual. New York: Cold Spring Harbor Laboratory Press; 1996.
- Babst M. A protein’s final ESCRT. Traffic. 2005;6:2–9. - PubMed
- Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell. 2002a;3:271–282. - PubMed
- Babst M., Katzmann D. J., Snyder W. B., Wendland B., Emr S. D. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 2002b;3:283–289. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases