Flexibility in a drug transport accessory protein: molecular dynamics simulations of MexA - PubMed (original) (raw)
Flexibility in a drug transport accessory protein: molecular dynamics simulations of MexA
Loredana Vaccaro et al. Biophys J. 2006.
Abstract
Drug resistance in gram-negative bacteria may be conferred via efflux through a tripartite complex of an inner membrane pump, an outer membrane pore, and a periplasmic adaptor protein. These are AcrB, TolC, and AcrA, respectively, in Escherichia coli. In Pseudomonas aerugonisa, their homologs are MexB, OprM, and MexA. Defining the interdomain dynamics of the adaptor protein is essential to understanding the mechanism of complex formation. Extended (25 ns) molecular dynamics simulations of MexA have been performed to determine such interdomain dynamics. Analysis of conformational drift demonstrates substantial motions of the three domains of MexA relative to one another. Principal components analysis reveals a hinge-bending motion and rotation of the alpha-helical hairpin relative to the other domains to be the two dominant motions. These two motions provide an element of considerable flexibility which is likely to be exploited in the adaptor function of MexA.
Figures
FIGURE 1
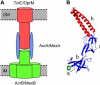
(A) Schematic diagram of the proposed transport complex formed by AcrB/MexB (green), AcrA/MexA (blue), and TolC/OprM (red) with the IM and the OM shown as gray bands. (B) The x-ray crystal structure of the MexA monomer from Pseudomonas aeruginosa. Three major domains can be distinguished: an _α_-helical hairpin (h, red); a _β_-domain (blue) made up of a lipoyl (l) and a _β_-barrel (b) subdomain; and a short _α_-helix (s, green).
FIGURE 2
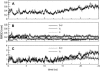
RMSD of the C_α_ atoms from their initial coordinates as a function of time, for simulation MD1. (A) RMSD for the whole protein. (B) RMSDs for the different domains (as defined in Fig. 1): the _α_-helical hairpin (h, black line), the _β_-domain (b + l, thick dark gray line), and the short α_-helix (s, thin pale gray line). (C) RMSD for the C_α atoms of the β_-domain (b + l, dark gray line) calculated while fitting onto the starting C_α coordinates of the _α_-helical hairpin (see text for details), and the RMSD for the α_-helical hairpin (h, black line) calculated while fitting onto the starting C_α coordinates of the _β_-domain.
FIGURE 3
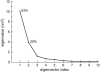
Eigenvalue spectra of the diagonalized covariance matrix, for simulation MD1. The first eigenvector contributes 53% to the total motion of the protein, whereas the second eigenvector contributes 20%.
FIGURE 4
Images of the motions corresponding to the first two eigenvectors for simulations MD1, MD2, and MD3. Each C_α_ atom has a cone attached pointing in the direction of motion described in the eigenvector for that atom. Thus, in simulations MD1, the first eigenvector (EV1) corresponds to a rotation of the small _α_-helix at the end of the _β_-barrel subdomain; and the second eigenvector (EV2) corresponds to a hinge-bending motion between the _α_-helical hairpin and the _β_-domain. The same pattern is seen in simulation MD3, whereas in MD2 the hinge-bending motion is EV1 and the rotation is EV2.
FIGURE 5
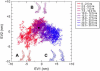
Projections of the trajectory of the MexA simulation onto the first (EV1) and second (EV2) eigenvectors. A gradient of colors from red to blue is used to track the protein over the 25 ns of simulation. The black circle indicates the projection of the starting structure onto the first and second eigenvectors. Two distinct conformational spaces are sampled during the simulation; the left-hand section of the plot (mostly in red) corresponds to hinge-bending of the two principal domains, whereas the right-hand section (mostly in blue) corresponds to rotation of the hairpin with respect to the _β_-strands. In red (A), we show the average structure over the first 5 ns, in purple (B) the average between 5 and 10 ns, and in blue (C) the average over the last 5 ns. In the first structure, an enlargement of the angle between the two domains is observed, corresponding to the hinge-bending motion, whereas the major difference between the blue structure and the x-ray structure lies in the bending of the hairpin.
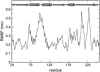
FIGURE 6
RMSF of C_α_ atoms with respect to their average position over the 25-ns simulation (MD1). A cartoon representation of the topology of the protein is shown above the graph.
FIGURE 7
Correlated motions in simulation MD1. (A) Motions with 50% correlation are indicated by lines connecting the correlated residues. Residues 106 and 213 are highlighted in green and blue, respectively. (B) Correlation matrix obtained by the covariance matrix; results are normalized and a gradient of colors from red (positive correlation) to blue (negative) is used. The results are normalized so that the extreme values of 1 and −1 correspond to complete correlation of motion at all the time, along the same direction, and along opposite directions, respectively. The box indicates the correlation for the _α_-helical hairpin. (C) Correlation of residue 213 (the blue sphere in A, located in the _β_-barrel domain) with the whole protein. The arrow indicates a positive correlation between this residue and the tip of the _α_-helical hairpin (residue 106).
Similar articles
- Structure of reconstituted bacterial membrane efflux pump by cryo-electron tomography.
Trépout S, Taveau JC, Benabdelhak H, Granier T, Ducruix A, Frangakis AS, Lambert O. Trépout S, et al. Biochim Biophys Acta. 2010 Oct;1798(10):1953-60. doi: 10.1016/j.bbamem.2010.06.019. Epub 2010 Jul 1. Biochim Biophys Acta. 2010. PMID: 20599691 - Assembly of the MexAB-OprM multidrug pump of Pseudomonas aeruginosa: component interactions defined by the study of pump mutant suppressors.
Nehme D, Poole K. Nehme D, et al. J Bacteriol. 2007 Sep;189(17):6118-27. doi: 10.1128/JB.00718-07. Epub 2007 Jun 22. J Bacteriol. 2007. PMID: 17586626 Free PMC article. - Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa.
Akama H, Matsuura T, Kashiwagi S, Yoneyama H, Narita S, Tsukihara T, Nakagawa A, Nakae T. Akama H, et al. J Biol Chem. 2004 Jun 18;279(25):25939-42. doi: 10.1074/jbc.C400164200. Epub 2004 Apr 26. J Biol Chem. 2004. PMID: 15117957 - The AcrB efflux pump: conformational cycling and peristalsis lead to multidrug resistance.
Seeger MA, Diederichs K, Eicher T, Brandstätter L, Schiefner A, Verrey F, Pos KM. Seeger MA, et al. Curr Drug Targets. 2008 Sep;9(9):729-49. doi: 10.2174/138945008785747789. Curr Drug Targets. 2008. PMID: 18781920 Review. - Three's company: component structures bring a closer view of tripartite drug efflux pumps.
Eswaran J, Koronakis E, Higgins MK, Hughes C, Koronakis V. Eswaran J, et al. Curr Opin Struct Biol. 2004 Dec;14(6):741-7. doi: 10.1016/j.sbi.2004.10.003. Curr Opin Struct Biol. 2004. PMID: 15582398 Review.
Cited by
- Structure of the periplasmic adaptor protein from a major facilitator superfamily (MFS) multidrug efflux pump.
Hinchliffe P, Greene NP, Paterson NG, Crow A, Hughes C, Koronakis V. Hinchliffe P, et al. FEBS Lett. 2014 Aug 25;588(17):3147-53. doi: 10.1016/j.febslet.2014.06.055. Epub 2014 Jul 1. FEBS Lett. 2014. PMID: 24996185 Free PMC article. - Computational study of correlated domain motions in the AcrB efflux transporter.
Schulz R, Vargiu AV, Ruggerone P, Kleinekathöfer U. Schulz R, et al. Biomed Res Int. 2015;2015:487298. doi: 10.1155/2015/487298. Epub 2015 Jan 5. Biomed Res Int. 2015. PMID: 25685792 Free PMC article. - The assembled structure of a complete tripartite bacterial multidrug efflux pump.
Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. Symmons MF, et al. Proc Natl Acad Sci U S A. 2009 Apr 28;106(17):7173-8. doi: 10.1073/pnas.0900693106. Epub 2009 Apr 2. Proc Natl Acad Sci U S A. 2009. PMID: 19342493 Free PMC article. - Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex.
Glavier M, Puvanendran D, Salvador D, Decossas M, Phan G, Garnier C, Frezza E, Cece Q, Schoehn G, Picard M, Taveau JC, Daury L, Broutin I, Lambert O. Glavier M, et al. Nat Commun. 2020 Oct 2;11(1):4948. doi: 10.1038/s41467-020-18770-5. Nat Commun. 2020. PMID: 33009415 Free PMC article. - A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps.
Lobedanz S, Bokma E, Symmons MF, Koronakis E, Hughes C, Koronakis V. Lobedanz S, et al. Proc Natl Acad Sci U S A. 2007 Mar 13;104(11):4612-7. doi: 10.1073/pnas.0610160104. Epub 2007 Mar 5. Proc Natl Acad Sci U S A. 2007. PMID: 17360572 Free PMC article.
References
- Lewis, K. 1994. Multidrug resistance pumps in bacteria: variations on a theme. Trends Biochem. Sci. 19:119–123. - PubMed
- Buchanan, S. K. 2001. Type I secretion and multidrug efflux: transport through the TolC channel-tunnel. Trends Biochem. Sci. 26:3–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous