Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells - PubMed (original) (raw)
Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells
Todd M Greco et al. Proc Natl Acad Sci U S A. 2006.
Abstract
S-nitrosylation, the selective modification of cysteine residues in proteins to form S-nitrosocysteine, is a major emerging mechanism by which nitric oxide acts as a signaling molecule. Even though nitric oxide is intimately involved in the regulation of vascular smooth muscle cell functions, the potential protein targets for nitric oxide modification as well as structural features that underlie the specificity of protein S-nitrosocysteine formation in these cells remain unknown. Therefore, we used a proteomic approach using selective peptide capturing and site-specific adduct mapping to identify the targets of S-nitrosylation in human aortic smooth muscle cells upon exposure to S-nitrosocysteine and propylamine propylamine NONOate. This strategy identified 20 unique S-nitrosocysteine-containing peptides belonging to 18 proteins including cytoskeletal proteins, chaperones, proteins of the translational machinery, vesicular transport, and signaling. Sequence analysis of the S-nitrosocysteine-containing peptides revealed the presence of acid/base motifs, as well as hydrophobic motifs surrounding the identified cysteine residues. High-resolution immunogold electron microscopy supported the cellular localization of several of these proteins. Interestingly, seven of the 18 proteins identified are localized within the ER/Golgi complex, suggesting a role for S-nitrosylation in membrane trafficking and ER stress response in vascular smooth muscle.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
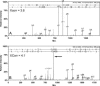
Fig. 1.
Evaluation of
sequest
peptide assignments. (A) An MS/MS spectrum (XCorr 3.6) assigned to an _S_-nitrosocysteine-containing peptide from 14-3-3 protein ζ that met all selection criteria and was accepted. (B) An MS/MS spectrum (Xcorr 4.1) assigned to a peptide from vimentin. Although this assignment passed the initial selection criteria, it was ultimately rejected because the top three most intense fragment peaks were not assigned (arrow). The evaluation of
sequest
peptide assignments was assessed by multiple selection criteria as follows: (i) Only peptide assignments that identified a biotin-HPDP derivitized cysteine (+428) included in the _y_- or _b_-ion series were considered. (ii) Each experimental condition was performed in quadruplicate, with peptide assignments evaluated if they appeared in at least three of the four independent replicates. (iii) Peptide assignments that passed these two selection filters were then evaluated by output scores assigned by
sequest
and were rejected if they did not meet specific threshold values as described in the Materials and Methods. (iv) If peptide assignments passed this scoring filter, the corresponding MS/MS spectra were manually reviewed. For an assignment to be accepted the MS/MS spectrum must have a continuous _b_- or _y_-ion series of at least five residues and the three most intense fragment peaks assigned to either an _a_-, _b_-, or _y_-ion, to an _a_-, _b_-, or _y_-ion resulting from a neutral loss of water or ammonia, or to a multiply protonated fragment ion. All review of peptide assignments and manual interpretation of MS/MS spectra were facilitated by
scaffold
, a proteome software package.
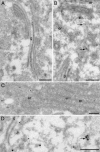
Fig. 2.
High-resolution immunoelectron microscopy. HASMC exposed to 100 μM CysNO for 20 min were fixed and processed for EM. Immunoreactivity for _S_-nitrosocysteine-containing proteins was visualized by 10-nm protein A gold particles. COP-1 immunoreactivity was visualized by 15-nm protein A gold particles. (A) Sections were treated with _para_-hydroxymercuricbenzoate to displace the _S_-nitrosocysteine adducts and then stained with monoclonal anti-_S_-nitrosocysteine antibody (26). (B) _S_-nitrosocysteine immunoreactivity (monoclonal antibody) was associated with endoplasmic reticulum (er) and small vesicular structures (arrows) in the vicinity of the Golgi complex (g). (C) A similar pattern of staining obtained with a polyclonal anti-_S_-nitrosocysteine antibody (asterisk indicates labeling of small vesicle). (D) Double labeling for _S_-nitrosocysteine (10-nm gold) and COP-1 (15-nm gold) showed localization on vesicular membrane profiles (arrow). (Scale bar, 200 nm.)
Fig. 3.
S-nitrosylation specificity motifs. (A) Sequence alignments of 18 _S_-nitrosocysteine-containing peptides identified from CysNO-treated HASMC comparing the occurrence of amino acids at positions flanking the modified cysteine. (B) Sequence alignments of 18 false positive peptides comparing the occurrence of amino acids at positions flanking the cysteine residue. (C) Kyte–Doolittle hydropathy plots from regions flanking the identified _S_-nitrosocysteine residue (arrow). The identified _S_-nitrosocysteine residues from T-complex protein 1, ζ subunit (Left), annexin A11 (Center), and elongation factor 1 A-1 (Right) were located within hydrophobic pockets. Hydropathy plots were constructed by using a window of 13 aa.
Similar articles
- Site specific identification of endogenous S-nitrosocysteine proteomes.
Doulias PT, Tenopoulou M, Raju K, Spruce LA, Seeholzer SH, Ischiropoulos H. Doulias PT, et al. J Proteomics. 2013 Oct 30;92:195-203. doi: 10.1016/j.jprot.2013.05.033. Epub 2013 Jun 5. J Proteomics. 2013. PMID: 23748021 Free PMC article. - Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation.
Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Doulias PT, et al. Proc Natl Acad Sci U S A. 2010 Sep 28;107(39):16958-63. doi: 10.1073/pnas.1008036107. Epub 2010 Sep 13. Proc Natl Acad Sci U S A. 2010. PMID: 20837516 Free PMC article. - Regulation of protein function and signaling by reversible cysteine S-nitrosylation.
Gould N, Doulias PT, Tenopoulou M, Raju K, Ischiropoulos H. Gould N, et al. J Biol Chem. 2013 Sep 13;288(37):26473-9. doi: 10.1074/jbc.R113.460261. Epub 2013 Jul 16. J Biol Chem. 2013. PMID: 23861393 Free PMC article. Review. - Mass spectrometry-based identification of S-nitrosocysteine in vivo using organic mercury assisted enrichment.
Doulias PT, Raju K, Greene JL, Tenopoulou M, Ischiropoulos H. Doulias PT, et al. Methods. 2013 Aug 1;62(2):165-70. doi: 10.1016/j.ymeth.2012.10.009. Epub 2012 Oct 29. Methods. 2013. PMID: 23116708 Free PMC article. Review. - Endogenous S-nitrosocysteine proteomic inventories identify a core of proteins in heart metabolic pathways.
Lau B, Fazelinia H, Mohanty I, Raimo S, Tenopoulou M, Doulias PT, Ischiropoulos H. Lau B, et al. Redox Biol. 2021 Nov;47:102153. doi: 10.1016/j.redox.2021.102153. Epub 2021 Oct 1. Redox Biol. 2021. PMID: 34610554 Free PMC article.
Cited by
- Site specific identification of endogenous S-nitrosocysteine proteomes.
Doulias PT, Tenopoulou M, Raju K, Spruce LA, Seeholzer SH, Ischiropoulos H. Doulias PT, et al. J Proteomics. 2013 Oct 30;92:195-203. doi: 10.1016/j.jprot.2013.05.033. Epub 2013 Jun 5. J Proteomics. 2013. PMID: 23748021 Free PMC article. - SILAM for quantitative proteomics of liver Akt1/PKBα after burn injury.
Lu XM, Tompkins RG, Fischman AJ. Lu XM, et al. Int J Mol Med. 2012 Mar;29(3):461-71. doi: 10.3892/ijmm.2011.861. Epub 2011 Dec 14. Int J Mol Med. 2012. PMID: 22179310 Free PMC article. - S-nitrosylation regulates nuclear translocation of chloride intracellular channel protein CLIC4.
Malik M, Shukla A, Amin P, Niedelman W, Lee J, Jividen K, Phang JM, Ding J, Suh KS, Curmi PM, Yuspa SH. Malik M, et al. J Biol Chem. 2010 Jul 30;285(31):23818-28. doi: 10.1074/jbc.M109.091611. Epub 2010 May 26. J Biol Chem. 2010. PMID: 20504765 Free PMC article. - S-sulfhydration/desulfhydration and S-nitrosylation/denitrosylation: a common paradigm for gasotransmitter signaling by H2S and NO.
Lu C, Kavalier A, Lukyanov E, Gross SS. Lu C, et al. Methods. 2013 Aug 1;62(2):177-81. doi: 10.1016/j.ymeth.2013.05.020. Epub 2013 Jun 27. Methods. 2013. PMID: 23811297 Free PMC article. Review. - Tetrahydrobiopterin modulates ubiquitin conjugation to UBC13/UBE2N and proteasome activity by S-nitrosation.
Bailey J, Davis S, Shaw A, Diotallevi M, Fischer R, Benson MA, Zhu H, Brown J, Bhattacharya S, Kessler BM, Channon KM, Crabtree MJ. Bailey J, et al. Sci Rep. 2018 Sep 25;8(1):14310. doi: 10.1038/s41598-018-32481-4. Sci Rep. 2018. PMID: 30254268 Free PMC article.
References
- Hess D. T., Matsumoto A., Kim S. O., Marshall H. E., Stamler J. S. Nat. Rev. Mol. Cell. Biol. 2005;6:150–166. - PubMed
- Hara M. R., Agrawal N., Kim S. F., Cascio M. B., Fujimuro M., Ozeki Y., Takahashi M., Cheah J. H., Tankou S. K., Hester L. D., et al. Nat. Cell Biol. 2005;7:665–674. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources