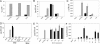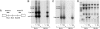Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection - PubMed (original) (raw)
Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection
Xiaolu Wu et al. Proc Natl Acad Sci U S A. 2006.
Abstract
The primate TRIM5alpha proteins have recently been defined as cellular restriction factors, preventing primate infection by retroviruses from different species. For instance, rhesus TRIM5alpha (rhTRIM5alpha) restricts infection by HIV-1. Virtually all TRIM5alpha proteins block the early replication of retroviruses by preventing the accumulation of reverse transcription products, but the underlying mechanism remains unclear. In this article, we find that disrupting proteasome function alters rhTRIM5alpha localization and allows the normal generation of HIV-1 late reverse transcription products, even though HIV-1 infection and the generation of nuclear 1-LTR and 2-LTR viral cDNA forms remain impaired. This finding suggests rhTRIM5alpha restricts HIV infection in two distinct phases: (i) altering the normal passage of the reverse-transcribing viral genome to the nucleus and (ii) targeting the reverse transcription complex to be disrupted by the proteasome. Because proteasome inhibitor blocks the second phase, accumulation of a nonfunctional viral DNA genome can be readily observed. Defining each phase may reveal HIV-1 targets for future antiviral therapy in which dual blockade may be equally as effective as naturally occurring rhTRIM5alpha protein in preventing HIV-1 infection in vivo.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
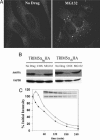
Fig. 1.
MG132 alters rhTRIM5α cytoplasmic localization. (A) rhTRIM5α.HA forms cytoplasmic bodies in stable HeLa cells (Left) that accumulate into larger bodies after 5 h of MG132 treatment (Right). (B) Western blot analysis reveals huTRIM5α.HA (Left) or rhTRIM5α.HA (Right) expression remains similar in HeLa cells after 6 h of MG132 treatment, despite declining with the translation inhibitor cyclohexamide (CHX). (C) Similar rhTRIM5α.HA stability without (Upper Inset; ♦) or with (Lower Inset; ▵) MG132 by pulse–chase analysis. Immunoprecipitated rhTRIM5α.HA bands are shown over time, with their percent intensity relative to the 0-min chase graphed.
Fig. 2.
MG132 relieves exogenous rhTRIM5α restriction of HIV-1 late RT products but not 2-LTR circles. (A_–_C) Untransfected (gray bars) or rhTRIM5α.HA stable HeLa (black bars) were pretreated 5 h with MG132, cyclohexamide (CHX), or nevirapine (Nev) and infected 14 h with VSV-g-pseudotyped R7-GFPHIV-1 plus drug. Cells were analyzed for infection at 48 h (A; percentage of cells expressing GFP shown) or viral late RT products (B), or 2-LTR circles (C) per cell at 14 h by real-time PCR. Error bars reflect the SD of triplicate values in a representative experiment. (D) A similar experiment except cells were pretreated 2 h with drugs, including BafA, before infection with either VSV-g-pseudotyped (VSV) or envelope minus (ΔEnv) R7-GFPHIV-1 plus drug. (E) A similar experiment except the MG132 pretreatment time was varied before infection with VSV-g-pseudotyped R7-GFPHIV-1. (F) Washing MG132 in or out at 4 or 22 h into infection reveals rhTRIM5α restricts late RT product formation early but not late in infection. MG132 added: ∗, pretreatment + 4 h infection; †, 4–22 h postinfection; ‡, 22–44 h postinfection.
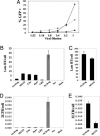
Fig. 3.
Proteasome inhibitors relieve endogenous rhTRIM5α restriction of HIV-1 late RT products. (A) Percentage of PRL (♦), HeLa (▴), or rhTRIM5α.HA HeLa (•) cells infected by VSV-g-pseudotyped R7-GFPHIV-1 dilutions, measured using the GFP reporter. (B_–_E) PRL cells (black bars) were preincubated either with MG132, cyclohexamide (CHX), or nevirapine (Nev) for 2 h or β-lactone or epoxomicin for 6 h and infected 18 h with 1:4-diluted (B and D) or undiluted (C and E) R7-GFPHIV-1 plus drug. Quantitative real-time PCR analysis shows proteasome inhibitors prevent endogenous rhTRIM5α restriction of late RT products (B) but not 2-LTR circles (D). HeLa cells infected with 1:4-diluted virus demonstrate unrestricted infection (gray bars).
Fig. 4.
MG132 allows extensive RT of HIV-1 cDNA despite exogenous rhTRIM5α. (A) Location of DNA digest sites and probes for Southern blot analysis of HIV-1 5′ (probe 1) and 3′ (probe 2) cDNA ends. (B_–_D) Southern blot analysis of uninfected (−) or R7-GFPHIV-1 infected HeLa or rhTRIM5α.HA stable HeLa (Rh HA) cells without (V) or with MG132 (MG). The same blot probed for 5′ (B) or 3′ (C) HIV-1 cDNA or GAPDH for DNA loading (D) is shown. Asterisks mark residual probe 2. Although late RT products with complete ends escape rhTRIM5α restriction via MG132, 1-LTR and 2-LTR circles remain impaired.
Similar articles
- Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5alpha.
Campbell EM, Perez O, Anderson JL, Hope TJ. Campbell EM, et al. J Cell Biol. 2008 Feb 11;180(3):549-61. doi: 10.1083/jcb.200706154. Epub 2008 Feb 4. J Cell Biol. 2008. PMID: 18250195 Free PMC article. - Recruitment and dynamics of proteasome association with rhTRIM5α cytoplasmic complexes during HIV-1 infection.
Danielson CM, Cianci GC, Hope TJ. Danielson CM, et al. Traffic. 2012 Sep;13(9):1206-17. doi: 10.1111/j.1600-0854.2012.01381.x. Epub 2012 Jun 19. Traffic. 2012. PMID: 22624877 Free PMC article. - TRIM5α-Mediated Ubiquitin Chain Conjugation Is Required for Inhibition of HIV-1 Reverse Transcription and Capsid Destabilization.
Campbell EM, Weingart J, Sette P, Opp S, Sastri J, O'Connor SK, Talley S, Diaz-Griffero F, Hirsch V, Bouamr F. Campbell EM, et al. J Virol. 2015 Dec 16;90(4):1849-57. doi: 10.1128/JVI.01948-15. Print 2016 Feb 15. J Virol. 2015. PMID: 26676782 Free PMC article. - Recent insights into the mechanism and consequences of TRIM5α retroviral restriction.
Sastri J, Campbell EM. Sastri J, et al. AIDS Res Hum Retroviruses. 2011 Mar;27(3):231-8. doi: 10.1089/AID.2010.0367. AIDS Res Hum Retroviruses. 2011. PMID: 21247355 Free PMC article. Review. - TRIM5alpha.
Song B. Song B. Curr Top Microbiol Immunol. 2009;339:47-66. doi: 10.1007/978-3-642-02175-6_3. Curr Top Microbiol Immunol. 2009. PMID: 20012523 Review.
Cited by
- TRIM22 inhibits influenza A virus infection by targeting the viral nucleoprotein for degradation.
Di Pietro A, Kajaste-Rudnitski A, Oteiza A, Nicora L, Towers GJ, Mechti N, Vicenzi E. Di Pietro A, et al. J Virol. 2013 Apr;87(8):4523-33. doi: 10.1128/JVI.02548-12. Epub 2013 Feb 13. J Virol. 2013. PMID: 23408607 Free PMC article. - HIV Restriction Factors and Mechanisms of Evasion.
Malim MH, Bieniasz PD. Malim MH, et al. Cold Spring Harb Perspect Med. 2012 May;2(5):a006940. doi: 10.1101/cshperspect.a006940. Cold Spring Harb Perspect Med. 2012. PMID: 22553496 Free PMC article. Review. - TRIM5 and the Regulation of HIV-1 Infectivity.
Luban J. Luban J. Mol Biol Int. 2012;2012:426840. doi: 10.1155/2012/426840. Epub 2012 May 30. Mol Biol Int. 2012. PMID: 22701176 Free PMC article. - The control of viral infection by tripartite motif proteins and cyclophilin A.
Towers GJ. Towers GJ. Retrovirology. 2007 Jun 12;4:40. doi: 10.1186/1742-4690-4-40. Retrovirology. 2007. PMID: 17565686 Free PMC article. Review. - Caging the beast: TRIM5α binding to the HIV-1 core.
Diaz-Griffero F. Diaz-Griffero F. Viruses. 2011 May;3(5):423-8. doi: 10.3390/v3050423. Epub 2011 Apr 27. Viruses. 2011. PMID: 21994740 Free PMC article.
References
- Bieniasz P. D. Nat. Immunol. 2004;5:1109–1115. - PubMed
- Kozak C. A., Chakraborti A. Virology. 1996;225:300–305. - PubMed
- Pincus T., Hartley J. W., Rowe W. P. Virology. 1975;65:333–342. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources