A functional network involved in the recycling of nucleocytoplasmic pre-60S factors - PubMed (original) (raw)
A functional network involved in the recycling of nucleocytoplasmic pre-60S factors
Alice Lebreton et al. J Cell Biol. 2006.
Abstract
Eukaryotic pre-ribosomes go through cytoplasmic maturation steps before entering translation. The nucleocytoplasmic proteins participating in these late stages of maturation are reimported to the nucleus. In this study, we describe a functional network focused on Rei1/Ybr267w, a strictly cytoplasmic pre-60S factor indirectly involved in nuclear 27S pre-ribosomal RNA processing. In the absence of Rei1, the nuclear import of at least three other pre-60S factors is impaired. The accumulation in the cytoplasm of a small complex formed by the association of Arx1 with a novel factor, Alb1/Yjl122w, inhibits the release of the putative antiassociation factor Tif6 from the premature large ribosomal subunits and its recycling to the nucleus. We propose a model in which Rei1 is a key factor for the coordinated dissociation and recycling of the last pre-60S factors before newly synthesized large ribosomal subunits enter translation.
Figures
Figure 1.
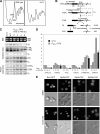
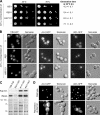
Rei1 is physically associated with nontranslating, Rpl24-containing complexes. (A) Rei1 is a two-hybrid prey of Rpl24b. YBR267W (REI1) was found as prey ORF in a genomic two-hybrid screen performed with RPL24B as bait. We represented the 393–amino acid–long protein and pointed out its putative motifs: three conserved C2H2 Zn fingers (dark gray) and the PFAM E-MAP-115 motif (stripes). Underneath, displayed as black rectangles, are the seven distinct DNA portions of REI1 that were found as preys. The minimal interacting domain is indicated in light gray. (B) Rei1-myc copurifies specifically with Rpl24b-TAP. TAPs were performed with strains expressing Rei1-13Myc together with Ssf1-TAP, Rlp24-TAP, Rpl24-TAP, or in a nontagged strain. The components of the purified complexes were separated by SDS-PAGE as shown on the Coomassie staining of the TEV eluate. Rei1-13Myc and Nog1 were detected in the TEV eluates by immunoblotting. As a loading control, Rei1-13Myc was detected in the input fractions (top). (C) Rei1 is associated with 60S particles. Whole cell extracts were prepared from a strain expressing Rpl24b-TAP and separated on a sucrose gradient by ultracentrifugation. Absorbance at 254 nm was measured, and 0.5-ml fractions of the gradient were collected. Peaks corresponding to the 40S, 60S, and 80S particles or to the polysomes are indicated. The proteins of each fraction were TCA precipitated and analyzed by Western blotting with antibodies directed against Rei1 or with PAP for the detection of the TAP fusion protein.
Figure 2.
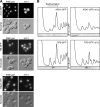
Absence of Rei1 affects the 60S maturation. (A) The rei1Δ strain displays an altered polysome profile. The disrupted strain and wild-type strain were grown for 16 h at 23°C in yeast extract–peptone–
d
-glucose (YPD). Whole cell extracts were separated on sucrose gradients as described in Fig. 1 C. In the rei1Δ strain (right), the position of half-mers is highlighted by asterisks. (B) Schematic representation of the maturation of the large subunit rRNA from 27SA2 to 5.8S and 25S. The positions of the oligonucleotide probes used in this study are indicated. (C) Effects of rei1Δ on pre-rRNA processing are similar to those of a TIF6 repression. Total RNAs were extracted from a rei1Δ or wild-type strain grown for 16 h at 23°C in YPD or from a PGAL1-TIF6 strain shifted to YPD from 0 to 6 h. 25S and 18S rRNAs were detected by ethidium bromide staining. 27SA2 and 27SB pre-rRNAs were detected by primer extension with probe CS10 (see Fig. 2 B). 7S pre-rRNA, 5.8S rRNA, and total 27S species were detected by Northern blotting with the CS3, CS5, and CS10 probes, respectively. (D) The absence of Rei1 or Tif6 correlates with a defect in the C1 + C2 rRNA processing step. Ratios were calculated either between two mature rRNA or pre-rRNA detected in the same samples on the same gel or after normalization with a control RNA (U2 small nuclear RNA) detected with oligonucleotide MFR457. RNA levels in the rei1Δ strain were standardized to the wild-type levels, indicated as a dotted line; for the TIF6 repression, the levels were indicated relative to those at time = 0. For rei1Δ, the calculation was performed on four distinct experiments; error bars represent SD. (E) Rei1 is not a shuttling factor. Strains producing Rei1-GFP, Rpl25-GFP, Rlp24-TAP, or Rpl24b-TAP were transformed with plasmids pAJ544 and pAJ368 overexpressing wild-type NMD3 or the dominant-negative nmd3Δ100 allele, respectively. The TAP fusion proteins were revealed by immunocytochemistry, and all samples were observed by epifluorescence microscopy.
Figure 3.
Rei1 is functionally linked to the shuttling factors Arx1 and Tif6. (A) In the absence of Rei1, Arx1 and Tif6 are redistributed to the cytoplasm. The localization of Arx1-GFP, Tif6-GFP, or Rlp24-TAP was observed after the growth of rei1Δ or wild-type strains for 8 h in minimal medium at 23°C. (B) Rei1 is required for the binding of Arx1 to pre-60S complexes. rei1Δ or wild-type strains expressing ARX1-GFP or TIF6-GFP were grown for 8 h at 23°C, and whole cell extracts were prepared. The extracts were separated on sucrose gradients and analyzed as described in Fig. 1 C. The immunoblots, which were performed using antibodies against GFP, are displayed below the corresponding profile.
Figure 4.
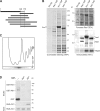
Yjl122w/Alb1 is a physical partner of Arx1. (A) Alb1 is a two-hybrid prey of Arx1. YJL122W (ALB1) was found as prey ORF in a genomic two-hybrid screen performed with ARX1 as bait. The results are presented as in Fig. 1 A. (B) Rei1, Arx1, and Alb1 are associated with the same complexes. Rei1-, Arx1-, and Alb1-TAP–associated complexes were TAP purified. The Coomassie staining is displayed on the left. The presence of Arx1, Rei1, or Rlp24 in the TEV eluates was assessed by immunoblotting with specific antibodies against each of these proteins, as shown on the right, with the corresponding Ponceau red staining. (C) Alb1-TAP is associated with 60S particles. Whole cell extracts were prepared from a strain expressing Alb1-TAP, separated on a sucrose gradient, and analyzed as described in Fig. 1 C. Alb1-TAP was detected by Western blotting with PAP. (D) Arx1 and Alb1 interact directly in vitro. GST, GST-Rei1, GST-Alb1, and His6-Arx1 fusion proteins were produced in Escherichia coli BL21. Crude extracts containing the GST-tagged baits were mixed with equal amounts of crude extracts containing the His6-Arx1 putative prey and were purified on glutathione–Sepharose beads. The input samples as well as the eluted proteins were separated on a 10% polyacrylamide-SDS gel. The bait proteins were detected in the eluted fractions by Western blotting with antibodies against GST (top). His6-Arx1 was detected with antibodies against Arx1 in the fractions where it is coeluted (middle). As a charge control, His6-Arx1 was also detected in the input (bottom).
Figure 5.
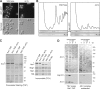
Alb1 is an additional shuttling pre-60S factor and target of Rei1. (A) In the absence of Rei1, Alb1 is blocked in the cytoplasm. The localization of Alb1-GFP was observed in rei1Δ or wild-type strains and grown for 8 h in minimal medium at 23°C. (B) Rei1 is required for the anchoring of Alb1 to pre-60S complexes. rei1Δ or wild-type strains expressing ALB1-GFP were grown for 16 h at 23°C, and whole cell extracts were prepared. The extracts were separated on sucrose gradients and analyzed as described in Fig. 1 C. The immunoblots, performed using antibodies against GFP or Arx1, are displayed below the corresponding profile. (C) Arx1 and Alb1 form a small nonribosomal complex in the absence of Rei1. Arx1- and Alb1-TAP–associated complexes were purified in wild-type or rei1Δ strains grown at 23°C. The Coomassie staining is shown on the left; the right panel displays the results of immunoblots using antibodies against Arx1, Nog1, Rei1, and Tif6 against the TEV eluates. Protein amounts were adjusted relative to Arx1. Positions of the tagged proteins are indicated by asterisks. (D) Arx1 and Kap121 cosediment with Alb1-TAP in rei1Δ strains. The TEV eluate from a purification of Alb1-TAP in the absence of Rei1 was further separated on sucrose gradients either directly (left) or after preincubation with calmodulin affinity resin (right; Alb1 complex subtracted). The components of the fractions were analyzed by immunoblotting using Arx1 and Kap121 antibodies after membrane staining by Ponceau red.
Figure 6.
Kap121 is involved in the Rei1-dependent nuclear import of pre-60S factors. (A) KAP121 is a high copy number suppressor of rei1Δ. A rei1Δ strain was transformed with high copy number plasmids carrying distinct DNA inserts (REI1, REH1, KAP121, or no insert) and plated in 10−1 dilution series on synthetic medium without uracil at 23 or 30°C for 3 and 2 d, respectively. Generation times at 23°C were calculated from growth curves performed in liquid synthetic medium without uracil over a period of 30 h. The confidence intervals were calculated for a probability of 95%. (B) Overexpression of KAP121 is sufficient to overcome the import defect in rei1Δ strains. rei1Δ strains expressing ARX1-GFP, ALB1-GFP, or TIF6-GFP were transformed with vectors overexpressing REI1, KAP121, or nothing. The localization of Arx1-, Alb1-, or Tif6-GFP was detected after growth for 8 h in minimal medium without uracil at 23°C. (C) Kap121 copurifies with Arx1-TAP and Alb1-TAP. Tif6-, Rei1-, Arx1-, and Alb1-TAP–associated complexes were purified by TAP. The presence of Kap121 or Rlp24 in the eluates was assessed by Western blotting with specific antibodies against each of these proteins. The Ponceau red staining is shown on the bottom. (D) In the absence of Kap121, Arx1-GFP is blocked in the cytoplasm. KAP121 was placed under the control of the PGAL1 promoter in strains expressing ARX1-GFP or ALB1-GFP. The localization of Arx1-GFP or Alb1-GFP was detected in this mutant or in the corresponding wild-type strain after growth in glucose- containing medium for 16 h.
Figure 7.
In the absence of Rei1, the cytoplasmic Arx1–Alb1 complex prevents the recycling of Tif6. (A) Disruptions of ARX1 or ALB1 compensate the cold-sensitive phenotype of a rei1Δ strain. Wild-type, rei1Δ, arx1Δ, or rei1Δ, arx1Δ strains were spotted on YPD medium in 10−1 dilution series (top); the same was performed for wild-type, rei1Δ, alb1Δ, or rei1Δ, alb1Δ strains (bottom). Cold-sensitive phenotypes were tested by incubating the plates for 3 d at 23°C. Generation times at 23°C were calculated from growth curves performed in liquid synthetic medium over a period of 30 h. The confidence intervals were calculated for a probability of 95%. (B) Disruptions of ARX1 or ALB1 compensate the Tif6 shuttling defect in a rei1Δ strain. The localization of Tif6-GFP was detected by epifluorescence microscopy in a wild-type, rei1Δ, and rei1Δ, alb1Δ or rei1Δ, arx1Δ double mutant strains.
Figure 8.
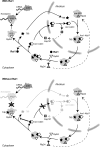
Model for the function of Rei1 at the exit of large ribosomal subunit biogenesis. Arx1, Alb1, and Tif6 are shuttling pre-60S factors that load on the precursors of the large ribosomal subunit in the nucleus and dissociate from the particles in the cytoplasm. At this last stage in biogenesis, the late ribosomal protein Rpl24 and the pre-60S factor Rei1 load on the particle. Rei1 triggers the Kap121-dependent recycling of Arx1 and Alb1 and subsequent dissociation and return of Tif6 to the nucleus. In the absence of Rei1, an Arx1, Alb1-containing subcomplex accumulates in the cytoplasm, which inhibits the dissociation of Tif6 from pre-60S particles. Thus, Tif6 antiassociation activity prevents the participation of novel large ribosomal subunits to translation initiation, and the lack of nuclear Tif6 results in pre-60S maturation defects.
Similar articles
- Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1.
Pertschy B, Saveanu C, Zisser G, Lebreton A, Tengg M, Jacquier A, Liebminger E, Nobis B, Kappel L, van der Klei I, Högenauer G, Fromont-Racine M, Bergler H. Pertschy B, et al. Mol Cell Biol. 2007 Oct;27(19):6581-92. doi: 10.1128/MCB.00668-07. Epub 2007 Jul 23. Mol Cell Biol. 2007. PMID: 17646390 Free PMC article. - Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae.
Hung NJ, Johnson AW. Hung NJ, et al. Mol Cell Biol. 2006 May;26(10):3718-27. doi: 10.1128/MCB.26.10.3718-3727.2006. Mol Cell Biol. 2006. PMID: 16648468 Free PMC article. - Factors affecting nuclear export of the 60S ribosomal subunit in vivo.
Stage-Zimmermann T, Schmidt U, Silver PA. Stage-Zimmermann T, et al. Mol Biol Cell. 2000 Nov;11(11):3777-89. doi: 10.1091/mbc.11.11.3777. Mol Biol Cell. 2000. PMID: 11071906 Free PMC article. - Nuclear export and cytoplasmic maturation of ribosomal subunits.
Zemp I, Kutay U. Zemp I, et al. FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review. - [Quality control of pre-ribosome coupled with ribosome biogenesis].
Matsuo Y. Matsuo Y. Seikagaku. 2014 Dec;86(6):793-7. Seikagaku. 2014. PMID: 25675820 Review. Japanese. No abstract available.
Cited by
- The NF45/NF90 Heterodimer Contributes to the Biogenesis of 60S Ribosomal Subunits and Influences Nucleolar Morphology.
Wandrey F, Montellese C, Koos K, Badertscher L, Bammert L, Cook AG, Zemp I, Horvath P, Kutay U. Wandrey F, et al. Mol Cell Biol. 2015 Oct;35(20):3491-503. doi: 10.1128/MCB.00306-15. Epub 2015 Aug 3. Mol Cell Biol. 2015. PMID: 26240280 Free PMC article. - Characterization of Saccharomyces cerevisiae Npa2p (Urb2p) reveals a low-molecular-mass complex containing Dbp6p, Npa1p (Urb1p), Nop8p, and Rsa3p involved in early steps of 60S ribosomal subunit biogenesis.
Rosado IV, Dez C, Lebaron S, Caizergues-Ferrer M, Henry Y, de la Cruz J. Rosado IV, et al. Mol Cell Biol. 2007 Feb;27(4):1207-21. doi: 10.1128/MCB.01523-06. Epub 2006 Dec 4. Mol Cell Biol. 2007. PMID: 17145778 Free PMC article. - Final pre-40S maturation depends on the functional integrity of the 60S subunit ribosomal protein L3.
García-Gómez JJ, Fernández-Pevida A, Lebaron S, Rosado IV, Tollervey D, Kressler D, de la Cruz J. García-Gómez JJ, et al. PLoS Genet. 2014 Mar 6;10(3):e1004205. doi: 10.1371/journal.pgen.1004205. eCollection 2014 Mar. PLoS Genet. 2014. PMID: 24603549 Free PMC article. - Dynamic association of human Ebp1 with the ribosome.
Bhaskar V, Desogus J, Graff-Meyer A, Schenk AD, Cavadini S, Chao JA. Bhaskar V, et al. RNA. 2021 Apr;27(4):411-419. doi: 10.1261/rna.077602.120. Epub 2021 Jan 21. RNA. 2021. PMID: 33479117 Free PMC article. - Multiple Puf proteins regulate the stability of ribosome biogenesis transcripts.
Fischer AD, Olivas WM. Fischer AD, et al. RNA Biol. 2018;15(9):1228-1243. doi: 10.1080/15476286.2018.1521211. Epub 2018 Sep 25. RNA Biol. 2018. PMID: 30251908 Free PMC article.
References
- Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 8:517–529. - PubMed
- Brachmann, C.B., A. Davies, G.J. Cost, E. Caputo, J. Li, P. Hieter, and J.D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132. - PubMed
- Dez, C., and D. Tollervey. 2004. Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 7:631–637. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases