Inhibitors of differentiation (ID1, ID2, ID3 and ID4) genes are neuronal targets of MeCP2 that are elevated in Rett syndrome - PubMed (original) (raw)
. 2006 Jun 15;15(12):2003-14.
doi: 10.1093/hmg/ddl124. Epub 2006 May 8.
Affiliations
- PMID: 16682435
- PMCID: PMC1931415
- DOI: 10.1093/hmg/ddl124
Inhibitors of differentiation (ID1, ID2, ID3 and ID4) genes are neuronal targets of MeCP2 that are elevated in Rett syndrome
Sailaja Peddada et al. Hum Mol Genet. 2006.
Abstract
Rett syndrome (RTT) is an X-linked dominant neurodevelopmental disorder caused by mutations in MECP2, encoding methyl-CpG-binding protein 2. MeCP2 is a transcriptional repressor elevated in mature neurons and is predicted to be required for neuronal maturation by regulating multiple target genes. Identifying primary gene targets in either Mecp2-deficient mice or human RTT brain has proven to be difficult, perhaps because of the transient requirement for MeCP2 during neuronal maturation. In order to experimentally control the timing of MeCP2 expression and deficiency during neuronal maturation, human SH-SY5Y cells undergoing mature neuronal differentiation were transfected with methylated MeCP2 oligonucleotide decoy to disrupt the binding of MeCP2 to endogenous targets. Genome-wide expression microarray analysis identified all four known members of the inhibitors of differentiation or inhibitors of DNA-binding (ID1, ID2, ID3 and ID4) subfamily of helix-loop-helix genes as novel neuronal targets of MeCP2. Chromatin immunoprecipitation analysis confirmed binding of MeCP2 near or within the promoters of ID1, ID2 and ID3, and quantitative RT-PCR confirmed increased expression of all four Id genes in Mecp2-deficient mouse brain. All four ID proteins were significantly increased in Mecp2-deficient mouse and human RTT brain using immunofluorescence and laser scanning cytometric analyses. Because of their involvement in cell differentiation and neural development, ID genes are ideal primary targets for MeCP2 regulation of neuronal maturation that may explain the molecular pathogenesis of RTT.
Figures
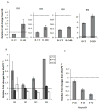
Figure 1. Quantitative RT-PCR results for ID1-4 in SH-SY5Y, _Mecp2_−/y mouse brain cDNA and NeuroD1, a downstream target of ID genes
A, B and C: Graphical representation of all four ID genes qPCR data reflect fold changes relative to the control (set to 1.0, indicated by hatched bar) following normalization of the data to GAPDH house keeping control using the comparative CT method. A) The qPCR data for ID1, ID2 and ID3 genes show a decreased expression with differentiation, in untransfected SH-SY5Y cells (D-UT) and increased expression with MeCP2 decoy (D-MD). qPCR data of ID4 shows an increased expression level following SH-SY5Y differentiation. ID4 was further increased in the D-MD transfected cells compared to 48 h untransfected (D-UT). Results shown represent mean ± SEM of three replicate experiments. B) The qRT-PCR was conducted in cDNA samples from _Mecp2_−/yand Mecp2+/ymice brains. The relative fold change differences for all four ID genes were higher in the Mecp2_−/y compared to Mecp2+/yat P28 time point, and reached significance by t-test for Id3 and Id4 (P < 0.05). No increase in cDNA was observed at later time points (P49 and P70). C) The qRT-PCR results on a downstream target of ID genes, NeuroD1, showed lower expression in all three postnatal time points (P28, P49 and P70) in Mecp2_−/y brain compared to the Mecp2+/y control brain, with significantly reduced expression at P49 and P70 by t-test (P ≤ 0.05). Each time point in bar graphs B and C represents mean ± SEM of three replicate experiments from two pairs of mice per time point.
Figure 2. Increased ID protein expression changes in _Mecp2-_deficient mice model detected by immunofluorescence and LSC
Immunofluorescence followed by analysis on LSC was performed on postnatal day 28, 5 μm sagittal brain sections of three pairs of _Mecp2_−/y and Mecp2+/y mice using antibodies recognizing ID1, ID2, ID3 and ID4. Representative LSC image showing protein expression differences in multiple regions of the brain of _Mecp2_−/y mice compared to Mecp2+/y controls for all four ID proteins. Each pixel represents individual nuclei colored blue (negative), green (low) or red (high) based on max pixel fluorescence histograms. For each section, the hemotoxylin and eosin (H&E) stained parallel slide was used to determine the tissue localization of cells and gating of individual brain regions for quantitative data shown in Table 3.
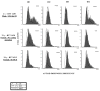
Figure 3. Increased ID protein expression changes in RTT cerebrum detected by immunofluorescence and LSC
Immunofluorescence using antibodies recognizing ID1-4 was performed on postmortem cerebral cortex samples from one male and two female RTT samples with and without MECP2 coding mutations (filled, grey histograms) and age-matched controls (open, black histogram) arranged on a tissue microarray. Expression of all four ID proteins was significantly increased in RTT brain samples compared to the controls. In contrast, histone H1 expression was not significantly changed (data not shown). Significance was determined using chi-square from two replicate experiments and the P values are denoted by asterix on the top right corner of each histogram. (* P ≤ 0.05, ** P ≤ 0.005, *** P ≤ 0.0005).
Figure 4. Chromatin Immunoprecipitation (ChIP) analysis of MeCP2 binding to ID1, ID2 and ID3 promoters
Representative gel of PCR products using primers specific to each of the ID gene promoter following ChIP with C-terminal MeCP2 reactive antibody. The total DNA isolated from chromatin prior to IP (Input) was used as a positive control. The ChIP results show binding of MeCP2 near or within the promoter of ID1, ID2 and ID3 in differentiated SH-SY5Y cells (D-UT) and control decoy transfected cells (D-CD). For ID4, binding of MeCP2 was not observed near or within the promoter region assayed. Additionally, the binding of MeCP2 to SNURF/SNRPN promoter is shown as a positive control for ChIP assay conditions.
Figure 5. Model for role of ID proteins in regulating neuronal maturational differentiation
In immature neurons with high expression of ID proteins, heterodimers of bHLH-ID prevent DNA binding and expression of differentiation associated genes like NEUROD1. Upon neuronal maturation, elevated levels of MeCP2 repress the expression of ID genes, resulting in functional bHLH-bHLH dimers that can bind to E-box DNA sequences and activate transcription of differentiation associated genes such as NEUROD1, ASCL1 and NEUROG1.
Similar articles
- Reciprocal co-regulation of EGR2 and MECP2 is disrupted in Rett syndrome and autism.
Swanberg SE, Nagarajan RP, Peddada S, Yasui DH, LaSalle JM. Swanberg SE, et al. Hum Mol Genet. 2009 Feb 1;18(3):525-34. doi: 10.1093/hmg/ddn380. Epub 2008 Nov 10. Hum Mol Genet. 2009. PMID: 19000991 Free PMC article. - Hormonal regulation and differential actions of the helix-loop-helix transcriptional inhibitors of differentiation (Id1, Id2, Id3, and Id4) in Sertoli cells.
Chaudhary J, Johnson J, Kim G, Skinner MK. Chaudhary J, et al. Endocrinology. 2001 May;142(5):1727-36. doi: 10.1210/endo.142.5.8134. Endocrinology. 2001. PMID: 11316735 - Mecp2 regulates neural cell differentiation by suppressing the Id1 to Her2 axis in zebrafish.
Gao H, Bu Y, Wu Q, Wang X, Chang N, Lei L, Chen S, Liu D, Zhu X, Hu K, Xiong JW. Gao H, et al. J Cell Sci. 2015 Jun 15;128(12):2340-50. doi: 10.1242/jcs.167874. Epub 2015 May 6. J Cell Sci. 2015. PMID: 25948585 - Regulation mechanism and research progress of MeCP2 in Rett syndrome.
Yang W, Pan H. Yang W, et al. Yi Chuan. 2014 Jul;36(7):625-30. doi: 10.3724/SP.J.1005.2014.0625. Yi Chuan. 2014. PMID: 25076025 Review. - MeCP2 expression and function during brain development: implications for Rett syndrome's pathogenesis and clinical evolution.
Kaufmann WE, Johnston MV, Blue ME. Kaufmann WE, et al. Brain Dev. 2005 Nov;27 Suppl 1:S77-S87. doi: 10.1016/j.braindev.2004.10.008. Epub 2005 Sep 22. Brain Dev. 2005. PMID: 16182491 Review.
Cited by
- Phosphorylation of distinct sites in MeCP2 modifies cofactor associations and the dynamics of transcriptional regulation.
Gonzales ML, Adams S, Dunaway KW, LaSalle JM. Gonzales ML, et al. Mol Cell Biol. 2012 Jul;32(14):2894-903. doi: 10.1128/MCB.06728-11. Epub 2012 May 21. Mol Cell Biol. 2012. PMID: 22615490 Free PMC article. - X-linked mental retardation and epigenetics.
Froyen G, Bauters M, Voet T, Marynen P. Froyen G, et al. J Cell Mol Med. 2006 Oct-Dec;10(4):808-25. doi: 10.1111/j.1582-4934.2006.tb00526.x. J Cell Mol Med. 2006. PMID: 17125586 Free PMC article. Review. - Downstream targets of methyl CpG binding protein 2 and their abnormal expression in the frontal cortex of the human Rett syndrome brain.
Gibson JH, Slobedman B, K N H, Williamson SL, Minchenko D, El-Osta A, Stern JL, Christodoulou J. Gibson JH, et al. BMC Neurosci. 2010 Apr 26;11:53. doi: 10.1186/1471-2202-11-53. BMC Neurosci. 2010. PMID: 20420693 Free PMC article. - A Small-Molecule Pan-Id Antagonist Inhibits Pathologic Ocular Neovascularization.
Wojnarowicz PM, Lima E Silva R, Ohnaka M, Lee SB, Chin Y, Kulukian A, Chang SH, Desai B, Garcia Escolano M, Shah R, Garcia-Cao M, Xu S, Kadam R, Goldgur Y, Miller MA, Ouerfelli O, Yang G, Arakawa T, Albanese SK, Garland WA, Stoller G, Chaudhary J, Norton L, Soni RK, Philip J, Hendrickson RC, Iavarone A, Dannenberg AJ, Chodera JD, Pavletich N, Lasorella A, Campochiaro PA, Benezra R. Wojnarowicz PM, et al. Cell Rep. 2019 Oct 1;29(1):62-75.e7. doi: 10.1016/j.celrep.2019.08.073. Cell Rep. 2019. PMID: 31577956 Free PMC article. - Evolving role of MeCP2 in Rett syndrome and autism.
LaSalle JM, Yasui DH. LaSalle JM, et al. Epigenomics. 2009 Oct;1(1):119-30. doi: 10.2217/epi.09.13. Epigenomics. 2009. PMID: 20473347 Free PMC article. Review.
References
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. - PubMed
- Zoghbi HY, Percy AK, Schultz RJ, Fill C. Patterns of X chromosome inactivation in the Rett syndrome. Brain Dev. 1990;12:131–135. - PubMed
- Armstrong DD. Review of Rett syndrome. J Neuropathol Exp Neurol. 1997;56:843–849. - PubMed
- Ellaway C, Christodoulou J. Rett syndrome: clinical update and review of recent genetic advances. J Paediat Child Health. 1999;35:419–426. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS041462/NS/NINDS NIH HHS/United States
- N01-HD-1-3138/HD/NICHD NIH HHS/United States
- MH/NS 31862/MH/NIMH NIH HHS/United States
- R01 HD041462/HD/NICHD NIH HHS/United States
- 1R01HD/NS41462/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases