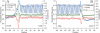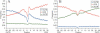A periodic pattern of mRNA secondary structure created by the genetic code - PubMed (original) (raw)
A periodic pattern of mRNA secondary structure created by the genetic code
Svetlana A Shabalina et al. Nucleic Acids Res. 2006.
Abstract
Single-stranded mRNA molecules form secondary structures through complementary self-interactions. Several hypotheses have been proposed on the relationship between the nucleotide sequence, encoded amino acid sequence and mRNA secondary structure. We performed the first transcriptome-wide in silico analysis of the human and mouse mRNA foldings and found a pronounced periodic pattern of nucleotide involvement in mRNA secondary structure. We show that this pattern is created by the structure of the genetic code, and the dinucleotide relative abundances are important for the maintenance of mRNA secondary structure. Although synonymous codon usage contributes to this pattern, it is intrinsic to the structure of the genetic code and manifests itself even in the absence of synonymous codon usage bias at the 4-fold degenerate sites. While all codon sites are important for the maintenance of mRNA secondary structure, degeneracy of the code allows regulation of stability and periodicity of mRNA secondary structure. We demonstrate that the third degenerate codon sites contribute most strongly to mRNA stability. These results convincingly support the hypothesis that redundancies in the genetic code allow transcripts to satisfy requirements for both protein structure and RNA structure. Our data show that selection may be operating on synonymous codons to maintain a more stable and ordered mRNA secondary structure, which is likely to be important for transcript stability and translation. We also demonstrate that functional domains of the mRNA [5'-untranslated region (5'-UTR), CDS and 3'-UTR] preferentially fold onto themselves, while the start codon and stop codon regions are characterized by relaxed secondary structures, which may facilitate initiation and termination of translation.
Figures
Figure 1
Profiles of nucleotide involvement in secondary structures, free energy of secondary structure formation and sequence conservation around the start codon (A) and the stop codon (B) in human mRNAs. Positions from −30 to −1 correspond to 5′-UTRs and positions from 1 to 60 correspond to CDSs (A). Positions from −60 to −1 correspond to CDSs and positions from 1 to 30 correspond to 3′-UTRs (B). Blue, sequence conservation in 6919 orthologous human and mouse mRNAs. Red, base paired nucleotides in 19 317 human mRNAs. Green, free Gibbs energy of base pairing in 19 317 human mRNAs.
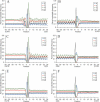
Figure 2
Profiles of nucleotide base pairing around the start codon (A, C and E) and the stop codon (B, D and F) for 19 317 human mRNAs (A and B), for sequences with randomly chosen synonymous codons (C and D), and sequences with randomly shuffled nucleotides and the same nucleotide composition as native mRNAs (E and F). Blue, guanine; red, cytosine; green, adenosine; orange, uridine.
Figure 3
The three phases of nucleotide base pairing in the mRNA CDS. The numbers denote codon sites.
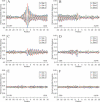
Figure 4
Profiles of base pairing for nucleotides around the start codon (A, C and E) and the stop codon (B, D and F) with different codon sites for 19 317 human mRNAs (A and B), for sequences with randomly chosen synonymous codons (C and D), and sequences with randomly shuffled nucleotides and the same nucleotide composition as native mRNAs (E and F). Nucleotides paired with codon sites 1, 2 and 3 are shown in blue, red and green, respectively.
Figure 5
Profiles of base pairing for nucleotides around the start codon (A) and the stop codon (B) with different mRNA structural domains. Blue, nucleotides paired with the 5′-UTRs; red, nucleotides paired with the CDSs; green, nucleotides paired with the 3′-UTRs. Data for 19 317 human mRNAs.
Similar articles
- Comparative analysis of orthologous eukaryotic mRNAs: potential hidden functional signals.
Shabalina SA, Ogurtsov AY, Rogozin IB, Koonin EV, Lipman DJ. Shabalina SA, et al. Nucleic Acids Res. 2004 Mar 18;32(5):1774-82. doi: 10.1093/nar/gkh313. Print 2004. Nucleic Acids Res. 2004. PMID: 15031317 Free PMC article. - RNA secondary structure profiling in zebrafish reveals unique regulatory features.
Kaushik K, Sivadas A, Vellarikkal SK, Verma A, Jayarajan R, Pandey S, Sethi T, Maiti S, Scaria V, Sivasubbu S. Kaushik K, et al. BMC Genomics. 2018 Feb 15;19(1):147. doi: 10.1186/s12864-018-4497-0. BMC Genomics. 2018. PMID: 29448945 Free PMC article. - Multiple RNA structures affect translation initiation and UGA redefinition efficiency during synthesis of selenoprotein P.
Mariotti M, Shetty S, Baird L, Wu S, Loughran G, Copeland PR, Atkins JF, Howard MT. Mariotti M, et al. Nucleic Acids Res. 2017 Dec 15;45(22):13004-13015. doi: 10.1093/nar/gkx982. Nucleic Acids Res. 2017. PMID: 29069514 Free PMC article. - A code within the genetic code: codon usage regulates co-translational protein folding.
Liu Y. Liu Y. Cell Commun Signal. 2020 Sep 9;18(1):145. doi: 10.1186/s12964-020-00642-6. Cell Commun Signal. 2020. PMID: 32907610 Free PMC article. Review. - A Code Within a Code: How Codons Fine-Tune Protein Folding in the Cell.
Komar AA. Komar AA. Biochemistry (Mosc). 2021 Aug;86(8):976-991. doi: 10.1134/S0006297921080083. Biochemistry (Mosc). 2021. PMID: 34488574 Free PMC article. Review.
Cited by
- A Parallel Study of mRNA and microRNA Profiling of Peripheral Blood in Young Adult Women.
Sredni ST, Gadd S, Jafari N, Huang CC. Sredni ST, et al. Front Genet. 2011 Jul 18;2:49. doi: 10.3389/fgene.2011.00049. eCollection 2011. Front Genet. 2011. PMID: 22303345 Free PMC article. - Attenuation of Human Respiratory Viruses by Synonymous Genome Recoding.
Le Nouën C, Collins PL, Buchholz UJ. Le Nouën C, et al. Front Immunol. 2019 Jun 4;10:1250. doi: 10.3389/fimmu.2019.01250. eCollection 2019. Front Immunol. 2019. PMID: 31231383 Free PMC article. Review. - Sequencing and comparative analysis of a conserved syntenic segment in the Solanaceae.
Wang Y, Diehl A, Wu F, Vrebalov J, Giovannoni J, Siepel A, Tanksley SD. Wang Y, et al. Genetics. 2008 Sep;180(1):391-408. doi: 10.1534/genetics.108.087981. Epub 2008 Aug 24. Genetics. 2008. PMID: 18723883 Free PMC article. - Widespread positive selection in synonymous sites of mammalian genes.
Resch AM, Carmel L, Mariño-Ramírez L, Ogurtsov AY, Shabalina SA, Rogozin IB, Koonin EV. Resch AM, et al. Mol Biol Evol. 2007 Aug;24(8):1821-31. doi: 10.1093/molbev/msm100. Epub 2007 May 23. Mol Biol Evol. 2007. PMID: 17522087 Free PMC article. - The Conservation and Function of RNA Secondary Structure in Plants.
Vandivier LE, Anderson SJ, Foley SW, Gregory BD. Vandivier LE, et al. Annu Rev Plant Biol. 2016 Apr 29;67:463-88. doi: 10.1146/annurev-arplant-043015-111754. Epub 2016 Feb 8. Annu Rev Plant Biol. 2016. PMID: 26865341 Free PMC article. Review.
References
- White H.B., III, Laux B.E., Dennis D. Messenger RNA structure: compatibility of hairpin loops with protein sequence. Science. 1972;175:1264–1266. - PubMed
- Ball L.A. Secondary structure and coding potential of the coat protein gene of bacteriophage MS2. Nature New Biol. 1973;242:44–45. - PubMed
- Fitch W.M. The large extent of putative secondary nucleic acid structure in random nucleotide sequences or amino acid derived messenger-RNA. J. Mol. Evol. 1974;3:279–291. - PubMed
- Lagunez-Otero J., Trifonov E.N. mRNA periodical infrastructure complementary to the proof-reading site in the ribosome. J. Biomol. Struct. Dyn. 1992;10:455–464. - PubMed
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources