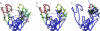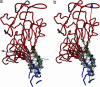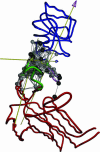Low-frequency normal modes that describe allosteric transitions in biological nanomachines are robust to sequence variations - PubMed (original) (raw)
Low-frequency normal modes that describe allosteric transitions in biological nanomachines are robust to sequence variations
Wenjun Zheng et al. Proc Natl Acad Sci U S A. 2006.
Abstract
By representing the high-resolution crystal structures of a number of enzymes using the elastic network model, it has been shown that only a few low-frequency normal modes are needed to describe the large-scale domain movements that are triggered by ligand binding. Here we explore a link between the nearly invariant nature of the modes that describe functional dynamics at the mesoscopic level and the large evolutionary sequence variations at the residue level. By using a structural perturbation method (SPM), which probes the residue-specific response to perturbations (or mutations), we identify a sparse network of strongly conserved residues that transmit allosteric signals in three structurally unrelated biological nanomachines, namely, DNA polymerase, myosin motor, and the Escherichia coli chaperonin. Based on the response of every mode to perturbations, which are generated by interchanging specific sequence pairs in a multiple sequence alignment, we show that the functionally relevant low-frequency modes are most robust to sequence variations. Our work shows that robustness of dynamical modes at the mesoscopic level is encoded in the structure through a sparse network of residues that transmit allosteric signals.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Fig. 1.
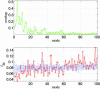
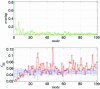
ENM and sequence analysis for Taq DNA polymerase’s open/closed transition (2KTQ → 3KTQ). (Upper) Overlap between each mode and the observed conformational changes for the lowest 100 modes. (Lower) The mode-dependent robustness (_f_δΕ) computed by the contact-based SPM for the lowest 100 modes, where the red curve (with circles) is for the original MSA and the blue curve (with error bars) represents the average (and standard deviation) of _f_δΕ distribution computed for 100 randomly permuted MSA.
Fig. 2.
Dynamical domain partitions, performed by using
dyndom
, for conformational changes described by the most robust modes, 4 (a), 7 (b), and 10 (c), of Taq DNA polymerase. The hinge regions are colored in green. The rotation axis for each pairwise interdomain motion is shown as an axis with an arrowhead; the color of the axis stem is set to be the same as the domain fixed for the structural alignment, whereas the color of the arrowhead is set to be the color of the moving domain. The hot-spot residues are shown in space-filled circles colored in gray. Residues in black have been experimentally identified to affect function (for residue numbers, see Table 2, which is published as supporting information on the PNAS web site).
Fig. 3.
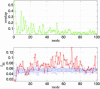
ENM and sequence analysis for Dictyostelium myosin (1VOM → 1MMA). The legend for the two graphs is the same as that for Fig. 1.
Fig. 4.
Dynamical domain partitions for conformational changes described by the most robust modes, 1 (a) and 2 (b), of Dictyostelium myosin. For the meaning of the relative domain motions given by the arrows, see the legend to Fig. 1. The hot-spot residues of modes 1 and 2 are shown in space-filled circles colored in gray. Residues in black have the same meaning as in Fig. 2 (for residue numbers, see Table 3, which is published as supporting information on the PNAS web site).
Fig. 5.
ENM and sequence analysis for E. coli GroEL (1AON_A → 1GRL). See the legend to Fig. 1 for a description.
Fig. 6.
Dynamical domain partitions for conformational changes described by the most robust mode 1 of E. coli GroEL. The description of the arrows indicating the relative motions of domains is the same as that given in the legend to Fig. 2. The hot-spot residues are shown in space-filled circles colored in gray. Residues in black have the same meaning as in Fig. 2 (for residue numbers, see Table 4, which is published as supporting information on the PNAS web site).
Similar articles
- Allosteric transitions in biological nanomachines are described by robust normal modes of elastic networks.
Zheng W, Brooks BR, Thirumalai D. Zheng W, et al. Curr Protein Pept Sci. 2009 Apr;10(2):128-32. doi: 10.2174/138920309787847608. Curr Protein Pept Sci. 2009. PMID: 19355980 Free PMC article. - Allostery wiring diagrams in the transitions that drive the GroEL reaction cycle.
Tehver R, Chen J, Thirumalai D. Tehver R, et al. J Mol Biol. 2009 Mar 27;387(2):390-406. doi: 10.1016/j.jmb.2008.12.032. Epub 2008 Dec 24. J Mol Biol. 2009. PMID: 19121324 - Allosteric transitions in the chaperonin GroEL are captured by a dominant normal mode that is most robust to sequence variations.
Zheng W, Brooks BR, Thirumalai D. Zheng W, et al. Biophys J. 2007 Oct 1;93(7):2289-99. doi: 10.1529/biophysj.107.105270. Epub 2007 Jun 8. Biophys J. 2007. PMID: 17557788 Free PMC article. - Signaling pathways regulating Dictyostelium myosin II.
De la Roche MA, Smith JL, Betapudi V, Egelhoff TT, Côté GP. De la Roche MA, et al. J Muscle Res Cell Motil. 2002;23(7-8):703-18. doi: 10.1023/a:1024467426244. J Muscle Res Cell Motil. 2002. PMID: 12952069 Review. - Signalling networks and dynamics of allosteric transitions in bacterial chaperonin GroEL: implications for iterative annealing of misfolded proteins.
Thirumalai D, Hyeon C. Thirumalai D, et al. Philos Trans R Soc Lond B Biol Sci. 2018 Jun 19;373(1749):20170182. doi: 10.1098/rstb.2017.0182. Philos Trans R Soc Lond B Biol Sci. 2018. PMID: 29735736 Free PMC article. Review.
Cited by
- Exploiting protein flexibility to predict the location of allosteric sites.
Panjkovich A, Daura X. Panjkovich A, et al. BMC Bioinformatics. 2012 Oct 25;13:273. doi: 10.1186/1471-2105-13-273. BMC Bioinformatics. 2012. PMID: 23095452 Free PMC article. - Coarse-grained models reveal functional dynamics--I. Elastic network models--theories, comparisons and perspectives.
Yang LW, Chng CP. Yang LW, et al. Bioinform Biol Insights. 2008 Mar 4;2:25-45. doi: 10.4137/bbi.s460. Bioinform Biol Insights. 2008. PMID: 19812764 Free PMC article. - A hierarchy of coupling free energies underlie the thermodynamic and functional architecture of protein structures.
Naganathan AN, Kannan A. Naganathan AN, et al. Curr Res Struct Biol. 2021 Oct 8;3:257-267. doi: 10.1016/j.crstbi.2021.09.003. eCollection 2021. Curr Res Struct Biol. 2021. PMID: 34704074 Free PMC article. Review. - Iterative cluster-NMA: A tool for generating conformational transitions in proteins.
Schuyler AD, Jernigan RL, Qasba PK, Ramakrishnan B, Chirikjian GS. Schuyler AD, et al. Proteins. 2009 Feb 15;74(3):760-76. doi: 10.1002/prot.22200. Proteins. 2009. PMID: 18712827 Free PMC article. - Differences in allosteric communication pipelines in the inactive and active states of a GPCR.
Bhattacharya S, Vaidehi N. Bhattacharya S, et al. Biophys J. 2014 Jul 15;107(2):422-434. doi: 10.1016/j.bpj.2014.06.015. Biophys J. 2014. PMID: 25028884 Free PMC article.
References
- Patel P. H., Loeb L. A. Nat. Struct. Biol. 2001;8:656–659. - PubMed
- Steitz T. J. Biol. Chem. 1999;274:17395–17398. - PubMed
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Nature. 1985;313:762. - PubMed
- Houdusse A., Sweeney H. Curr. Opin. Struct. Biol. 2001;11:182–194. - PubMed
- Xu Z., Horwich A. L., Sigler P. B. Nature. 1997;388:741–750. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources