Clusters of internally primed transcripts reveal novel long noncoding RNAs - PubMed (original) (raw)
Comparative Study
doi: 10.1371/journal.pgen.0020037. Epub 2006 Apr 28.
Ken C Pang, Noriko Ninomiya, Shiro Fukuda, Martin C Frith, Carol Bult, Chikatoshi Kai, Jun Kawai, Piero Carninci, Yoshihide Hayashizaki, John S Mattick, Harukazu Suzuki
Affiliations
- PMID: 16683026
- PMCID: PMC1449886
- DOI: 10.1371/journal.pgen.0020037
Comparative Study
Clusters of internally primed transcripts reveal novel long noncoding RNAs
Masaaki Furuno et al. PLoS Genet. 2006 Apr.
Abstract
Non-protein-coding RNAs (ncRNAs) are increasingly being recognized as having important regulatory roles. Although much recent attention has focused on tiny 22- to 25-nucleotide microRNAs, several functional ncRNAs are orders of magnitude larger in size. Examples of such macro ncRNAs include Xist and Air, which in mouse are 18 and 108 kilobases (Kb), respectively. We surveyed the 102,801 FANTOM3 mouse cDNA clones and found that Air and Xist were present not as single, full-length transcripts but as a cluster of multiple, shorter cDNAs, which were unspliced, had little coding potential, and were most likely primed from internal adenine-rich regions within longer parental transcripts. We therefore conducted a genome-wide search for regional clusters of such cDNAs to find novel macro ncRNA candidates. Sixty-six regions were identified, each of which mapped outside known protein-coding loci and which had a mean length of 92 Kb. We detected several known long ncRNAs within these regions, supporting the basic rationale of our approach. In silico analysis showed that many regions had evidence of imprinting and/or antisense transcription. These regions were significantly associated with microRNAs and transcripts from the central nervous system. We selected eight novel regions for experimental validation by northern blot and RT-PCR and found that the majority represent previously unrecognized noncoding transcripts that are at least 10 Kb in size and predominantly localized in the nucleus. Taken together, the data not only identify multiple new ncRNAs but also suggest the existence of many more macro ncRNAs like Xist and Air.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
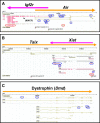
Figure 1. Snapshots of the GEV Showing Transcription
(A) The Air/Igf2r locus (Chromosome 17: 12,091,531–12,258,195). (B) The Xist/Tsix locus (X chromosome: 94,835,096–94,888,536). (C) The dystrophin (Dmd) locus (X chromosome: 76,500,000–76,754,601). For the transcripts, cDNA sequences from the RIKEN and public databases are shown, and are colored in brown and purple depending upon their chromosomal strand of origin. Predicted genes from Ensembl, NCBI, and RefSeq databases are shown in gray. CpG islands as defined by the UCSC Genome Browser are shown. Blue circles indicate unspliced, noncoding RIKEN cDNAs with adjunct adenine-rich regions. Red circles indicate RIKEN imprinted cDNA candidates [38].
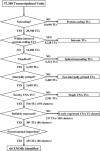
Figure 2. Discovery Pipeline for ENORs
FANTOM and public transcripts were clustered into 37,348 TUs by grouping any two or more transcripts that shared genomic coordinates. Then, the following procedures were applied. (1) Protein-coding TUs were excluded by removing any whose transcripts had an open reading frame of either 150 amino acids or more (RIKEN/MGC cDNAs) or one amino acid or more (non-RIKEN/MGC cDNAs). (2) TUs wholly encompassed within introns of protein-coding TUs were excluded to avoid possible pre-mRNA intronic transcripts. (3) Intron-containing TUs were excluded to select for unspliced transcripts. (4) TUs lacking adjunct adenine-rich regions or containing polyA signals were excluded to select for internally primed transcripts. (5) Remaining UNA TUs that mapped within 100 Kb of one another on the mouse genome (mm5) were clustered together, provided they did not overlap the genomic coordinates of a protein-coding TU/NCBI RefSeq/Ensembl gene model with a CDS of 150 amino acids or more or a noncoding TU with a polyA signal within 100 bp of the 3′ end and without an adjunct adenine-rich region. (6) Reliably expressed UNA TU clusters were selected by identifying those with at least ten supporting ESTs. (7) Selected UNA TU clusters were then manually screened and separated based upon evidence of possible internal transcription state sites (based upon CpG islands, CAGE tags, and EST clusters), resulting in the identification of 66 ENORs.
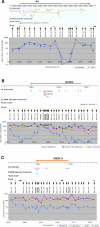
Figure 3. ENOR Tissue Expression
Tissue expression information for individual ENORs was obtained using publicly available GNF Gene Expression Atlas data. GNF probes that overlapped ENORs were identified, and the corresponding relative expression ratios for 61 tissues were hierarchically clustered. Red squares indicate high expression, black squares indicate low expression, and grey squares indicate where expression was not reliably detected (based upon Affymetrix MAS5 absent/present calls). med. olfactory epi., medial olfactory epithelium.
Figure 4. qRT-PCR Analysis
Analysis of (A) Air, (B) ENOR28, and (C) ENOR31 loci. Above in each panel, screen shots of the GEV featuring the loci around Air, ENOR28, and ENOR31 are shown. The orange bars indicate the regions for Air, ENOR28, and ENOR31. cDNA sequences from the RIKEN and public databases are shown. Sequences mapped on the plus strand and minus strand are brown and purple, respectively. Predicted genes from Ensembl, NCBI, and RefSeq databases are shown in gray. For RIKEN imprinted transcripts, imprinted cDNA candidates identified previously [38] are shown. CpG islands as defined by the UCSC Genome Browser are shown. Positions of primer pairs are marked by small vertical arrows. Below in each panel, qRT-PCR results for midbrain, hippocampus, thalamus, striatum, and testis using the corresponding primer pairs are shown.
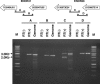
Figure 5. Presence of Transcription between Adjacent cDNAs
PCR was carried out with and without reverse transcription (RT[+] and RT[−], respectively) using midbrain total RNA and the corresponding primer pairs (see Table S3). PCR using genomic DNA was also carried out as a control. A DNA ladder (Promega;
) was used as a size marker. The amplified fragments were confirmed as the expected ones by analyzing digestion pattern using several restriction enzymes. The lower band, observed in the RT(+) lane of the amplified fragment C, seems to be nonspecific, because it was amplified using only the right primer and because it showed a digestion pattern with restriction enzymes quite different from that of the upper band and the band of the genomic DNA (unpublished data).
Figure 6. Northern Blot Analysis of ENOR Transcripts
Mouse whole brain total RNA (10 μg/lane) was used for the analysis except for ENOR2 and ENOR61, where mouse thymus total RNA was used. DNA fragments without any predicted repeated sequences were PCR-amplified from cDNAs in ENORs (Table S3), labeled with 32P-dCTP (Amersham Biosciences), and then used as probes. RNA size was estimated with an RNA ladder (Invitrogen). ENORs are listed in increasing order based on the estimated length of each region.
Figure 7. Localization of ENOR Transcripts
qRT-PCR was carried out using total and cytoplasmic RNA from mouse whole brain and the corresponding primer pairs (Table S3). ENORs are listed in increasing order based on the estimated length of each region. Apart from the results shown, we also examined the localization of other mRNAs (β-actin and GAPDH) and additional regions of Rian and other ENORs, and these results were consistent with the rest (unpublished data).
Similar articles
- Critical evaluation of the FANTOM3 non-coding RNA transcripts.
Nordström KJ, Mirza MA, Almén MS, Gloriam DE, Fredriksson R, Schiöth HB. Nordström KJ, et al. Genomics. 2009 Sep;94(3):169-76. doi: 10.1016/j.ygeno.2009.05.012. Epub 2009 Jun 6. Genomics. 2009. PMID: 19505569 - Identification of putative noncoding RNAs among the RIKEN mouse full-length cDNA collection.
Numata K, Kanai A, Saito R, Kondo S, Adachi J, Wilming LG, Hume DA, Hayashizaki Y, Tomita M; RIKEN GER Group; GSL Members. Numata K, et al. Genome Res. 2003 Jun;13(6B):1301-6. doi: 10.1101/gr.1011603. Genome Res. 2003. PMID: 12819127 Free PMC article. - Silencing by imprinted noncoding RNAs: is transcription the answer?
Pauler FM, Koerner MV, Barlow DP. Pauler FM, et al. Trends Genet. 2007 Jun;23(6):284-92. doi: 10.1016/j.tig.2007.03.018. Epub 2007 Apr 18. Trends Genet. 2007. PMID: 17445943 Free PMC article. Review. - Finding noncoding RNA transcripts from low abundance expressed sequence tags.
Xue C, Li F, Li F. Xue C, et al. Cell Res. 2008 Jun;18(6):695-700. doi: 10.1038/cr.2008.59. Cell Res. 2008. PMID: 18504459 - [Non-coding RNA - from useless to essential].
[No authors listed] [No authors listed] Cas Lek Cesk. 2016 Winter;155(7):370-376. Cas Lek Cesk. 2016. PMID: 27990832 Review. Czech.
Cited by
- Evaluation of immune sensor responses to a viral small noncoding RNA.
Kara M, Tibbetts SA. Kara M, et al. Front Cell Infect Microbiol. 2024 Oct 8;14:1459256. doi: 10.3389/fcimb.2024.1459256. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39450336 Free PMC article. - Tumor diagnosis using carbon-based quantum dots: Detection based on the hallmarks of cancer.
Wang H, Yang S, Chen L, Li Y, He P, Wang G, Dong H, Ma P, Ding G. Wang H, et al. Bioact Mater. 2023 Nov 15;33:174-222. doi: 10.1016/j.bioactmat.2023.10.004. eCollection 2024 Mar. Bioact Mater. 2023. PMID: 38034499 Free PMC article. Review. - M6A-mediated-upregulation of lncRNA BLACAT3 promotes bladder cancer angiogenesis and hematogenous metastasis through YBX3 nuclear shuttling and enhancing NCF2 transcription.
Xie J, Zhang H, Wang K, Ni J, Ma X, Khoury CJ, Prifti V, Hoard B, Cerenzia EG, Yin L, Zhang H, Wang R, Zhuo D, Mao W, Peng B. Xie J, et al. Oncogene. 2023 Sep;42(40):2956-2970. doi: 10.1038/s41388-023-02814-3. Epub 2023 Aug 23. Oncogene. 2023. PMID: 37612524 Free PMC article. - Long non-coding RNA BRE-AS1 inhibits proliferation, migration and invasion of clear cell renal cell carcinoma by downregulating miR-106b-5p.
Zou G, Guo L, Shen S, Song Z, Ouyang Z, Yu Y. Zou G, et al. Histol Histopathol. 2024 Jun;39(6):729-737. doi: 10.14670/HH-18-653. Epub 2023 Jul 25. Histol Histopathol. 2024. PMID: 37530129 - Host gene and its guest: short story about relation of long-noncoding MIR31HG transcript and microRNA miR-31.
Kolenda T, Paszkowska A, Braska A, Kozłowska-Masłoń J, Guglas K, Poter P, Wojtczak P, Bliźniak R, Lamperska K, Teresiak A. Kolenda T, et al. Rep Pract Oncol Radiother. 2023 Apr 6;28(1):114-134. doi: 10.5603/RPOR.a2023.0006. eCollection 2023. Rep Pract Oncol Radiother. 2023. PMID: 37122913 Free PMC article. Review.
References
- Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14((Suppl 1)):R121–R132. - PubMed
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. - PubMed
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. - PubMed
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources