Complex Loci in human and mouse genomes - PubMed (original) (raw)
Comparative Study
doi: 10.1371/journal.pgen.0020047. Epub 2006 Apr 28.
Harukazu Suzuki, Noriko Ninomiya, Altuna Akalin, Luca Sessa, Giovanni Lavorgna, Alessandro Brozzi, Lucilla Luzi, Sin Lam Tan, Liang Yang, Galih Kunarso, Edwin Lian-Chong Ng, Serge Batalov, Claes Wahlestedt, Chikatoshi Kai, Jun Kawai, Piero Carninci, Yoshihide Hayashizaki, Christine Wells, Vladimir B Bajic, Valerio Orlando, James F Reid, Boris Lenhard, Leonard Lipovich
Affiliations
- PMID: 16683030
- PMCID: PMC1449890
- DOI: 10.1371/journal.pgen.0020047
Comparative Study
Complex Loci in human and mouse genomes
Pär G Engström et al. PLoS Genet. 2006 Apr.
Abstract
Mammalian genomes harbor a larger than expected number of complex loci, in which multiple genes are coupled by shared transcribed regions in antisense orientation and/or by bidirectional core promoters. To determine the incidence, functional significance, and evolutionary context of mammalian complex loci, we identified and characterized 5,248 cis-antisense pairs, 1,638 bidirectional promoters, and 1,153 chains of multiple cis-antisense and/or bidirectionally promoted pairs from 36,606 mouse transcriptional units (TUs), along with 6,141 cis-antisense pairs, 2,113 bidirectional promoters, and 1,480 chains from 42,887 human TUs. In both human and mouse, 25% of TUs resided in cis-antisense pairs, only 17% of which were conserved between the two organisms, indicating frequent species specificity of antisense gene arrangements. A sampling approach indicated that over 40% of all TUs might actually be in cis-antisense pairs, and that only a minority of these arrangements are likely to be conserved between human and mouse. Bidirectional promoters were characterized by variable transcriptional start sites and an identifiable midpoint at which overall sequence composition changed strand and the direction of transcriptional initiation switched. In microarray data covering a wide range of mouse tissues, genes in cis-antisense and bidirectionally promoted arrangement showed a higher probability of being coordinately expressed than random pairs of genes. In a case study on homeotic loci, we observed extensive transcription of nonconserved sequences on the noncoding strand, implying that the presence rather than the sequence of these transcripts is of functional importance. Complex loci are ubiquitous, host numerous nonconserved gene structures and lineage-specific exonification events, and may have a cis-regulatory impact on the member genes.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
Figure 1. TU Pairs Searched For
We defined a _cis–_antisense pair as two oppositely transcribed TUs that share at least 20 bp of exon sequence, a non-exon-overlapping antisense pair as two oppositely transcribed TUs that overlap by at least 20 bp, but not within exons, and a bidirectionally promoted pair as two divergently transcribed TUs that overlap by less than 20 bp and are less than 1,000 bp apart.
Figure 2. Validation of the Expression of Randomly Selected _cis–_Antisense Pairs by RT-PCR
To confirm the expression of complementary transcripts, we performed orientation-specific RT-PCR as described previously [12,24]. Primers were designed to amplify regions of exon overlap. For each candidate or control, four RT-PCR reactions (corresponding to the four lanes in each gel image) were carried out using adult mouse brain RNA as template. Orientation specificity was achieved by restricting which primers were present during reverse transcription single-strand synthesis: no primer (first lane), only sense primer (second lane), only antisense primer (third lane), and both sense and antisense primers (fourth lane). In all reactions, both primers were present during the subsequent PCR reactions. For candidates, sense and antisense primers were designed with respect to the genomic plus strand. For controls, primers were designed with respect to the control transcript. Out of five highly expressed control genes with no evidence of antisense transcription in sequence databases, we detected antisense transcription for one (Rps27). We reproducibly observed evidence of anti-Rps27 transcripts using two different primer pairs (unpublished data). We tested 20 _cis–_antisense pairs from our computationally constructed dataset and detected expression of both strands for 16 (underlined). For one additional _cis–_antisense pair (number 11), the result was ambiguous because of the presence of many bands of unexpected size. The 20 _cis–_antisense pairs were selected at random from the mouse dataset, with the requirements that exon overlaps be at least 200 bp (to allow amplicons of at least 100 bp) and that there be at least one cDNA or EST from adult brain supporting the exon overlap on each strand.
Figure 3. Estimating the Extent and Conservation of Antisense Transcription
(A and B) Estimation of proportion of TUs involved in _cis–_antisense pairs. Open circles indicate the fraction of all human TUs on the plus strand (A) and all mouse TUs on the plus strand (B) that were found to be involved in _cis–_antisense pairs when the minus-strand TUs were recomputed starting from random transcript sequence samples of different sizes. Filled circles represent the full datasets based on all available transcript sequences. The saturation curves (see Equation 1) indicated by the lines fit almost perfectly to the sampled data. Fitted human and mouse saturation curves approach 0.45 and 0.43, respectively, as the number of transcript sequences increases, indicating that more than 40% of all TUs might be involved in _cis–_antisense pairs. Similar estimates were obtained by other sampling approaches (Figure S3). (C) Estimation of the proportion of human _cis–_antisense pairs that are conserved in mouse. Open circles indicate the proportion of human _cis–_antisense pairs found to be conserved in mouse when the full human dataset was compared to mouse datasets recomputed from random mouse transcript sequence samples of different sizes. The same type of saturation curve as in (A) was fitted to the data. Here, a model with c = 1 (i.e., hyperbolic saturation) was preferable as it provided an equally good fit while being simpler. The fitted curve approaches 0.25 as the number of mappings grows, indicating that about 25% of human _cis–_antisense pairs are conserved in mouse.
Figure 4. Positional Equivalents
(A) Schematic depiction of positional equivalents. By positional equivalents (red arrows), we mean mouse and human TUs that are at genomically equivalent locations relative to well annotated genes at orthologous loci (blue arrows), but that do not share sequence similarity. (B) Positional equivalents divergently transcribed with the putative tumor suppressor RNH1 [48]. Two transcript isoforms of RNH1 are shown for both mouse (top) and human (bottom). A mouse TU supported by cDNA AK020472 shares a putative bidirectional promoter with Rnh1. The human equivalent (cDNA AK095144) is head-to-head _cis–_antisense to RNH1. Regions with gray background are within a BLASTZ net alignment of the two genomes. For Rnh1 and RNH1, protein-coding sequence is indicated in dark blue and UTRs in light blue. The positional equivalents lack sequence conservation, assessed by BLASTZ net coverage and BL2SEQ alignment of transcripts, demonstrate gene structure differences, and contain lineage-specific repeats (indicated in black).
Figure 5. TSS Variability at the Ddx49/Cope Bidirectional Promoter in Mouse
(A) The charts show the distribution of CAGE tag 5′-ends over the first five exons of each of the two genes Ddx49 and Cope, and over their intergenic region. CAGE tag mappings indicate that transcription of Cope can start within two wide regions in the first exon of the gene. The initial part of this first exon (hatched) has support from several ESTs, but no cDNA sequences. The three large TCs at the Ddx49/Cope locus span 79, 114, and 150 bp, indicating great variability of transcriptional initiation within each cluster. To confirm the existence of such variability by qRT-PCR, primers (connected boxes) were designed to measure expression of selected regions of the Ddx49 (primer pairs A1–A4) and Cope (primer pairs B1–B5) transcripts. (B) Detailed view of CAGE tag frequencies and primer locations over the three transcription initiation regions indicated by CAGE tags. Gray lines show cumulative CAGE tag frequencies. (C) Expression levels of different regions of the Ddx49 and Cope transcripts in adult brain RNA as measured by qRT-PCR. Primer pairs A1 and A2 confirmed low level of expression of the longest Ddx49 transcripts indicated by CAGE (copy numbers in 12.5 ng of total RNA were 3.2 [standard deviation = 1.1] and 5.1 [standard deviation = 3.0] for A1 and A2, respectively). Primer pair B1 confirmed transcription of Cope from upstream of the canonical initiation region. Primer pairs B2–B4 supported variability of transcriptional initiation within the canonical region.
Figure 6. Landmark Sequence Composition of Bidirectional Promoters
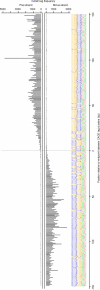
We defined the midpoint of a bidirectional promoter as the midpoint between the most 5′ TSS in each of the two divergently oriented TCs defining the bidirectional promoter. Sequences corresponding to the region spanned by the TCs were extracted from the genomic plus strand. All bidirectional promoter sequences were aligned at their midpoint and the logo created with WebLogo [49]. The logo displays the four nucleotides ranked by their frequency at each position, so that more common nucleotides appear above less common ones. The charts above the logo show the distribution of CAGE tag 5′-ends mapping to the plus strand (upper chart) and minus strand (lower chart) around bidirectional promoter midpoints. The CAGE tag distribution was computed as the sum of tag counts at each position over all bidirectional promoters. The peak of nearly 5,000 tags on the plus strand is due to the Rps2 gene, which appears to be most highly expressed from a single TSS.
Figure 7. Members of _cis–_Antisense Pairs Have Positively Correlated Expression Profiles More Often than Expected by Chance
Out of 242 murine _cis–_antisense pairs with expression data for 61 tissues, 17 showed significant positive correlation across the entire set of tissues after correction for multiple testing, and no pairs showed significant negative correlation (red squares). The same test was applied to 10,000 sets of 242 random TU pairs (box plots, with circles indicating outliers), demonstrating that members of _cis–_antisense pairs have positively correlated expression profiles more often than expected by chance.
Figure 8. A Five-TU Chain on Mouse Chromosome 15
TUs on the genomic plus and minus strands are shown in dark gray and light gray, respectively (boxes represent exons). CpG islands are shown as black boxes. From left to right, the chain contains a member of the aminoacyl tRNA transferase class II family (D330001F17Rik), which has two _cis–_antisense transcripts: fully overlapping (cDNA AK034666) and convergent (Bop1). The latter encodes a ribosome biogenesis protein and shares a CpG-island bidirectional promoter with the heat-shock-induced transcription factor 1 gene (Hsf1). Hsf1, in turn, is convergently _cis–_antisense to the diacylglycerol O-acyltransferase 1 gene (Dgat1).
Figure 9. Dispersed Human and Mouse Homeotic Loci at Which ESTs Were Detected on the Opposite Strand from the Homeotic Gene
Loci with opposite-strand ESTs in both genomes are listed in the center box.
Similar articles
- Abundant novel transcriptional units and unconventional gene pairs on human chromosome 22.
Lipovich L, King MC. Lipovich L, et al. Genome Res. 2006 Jan;16(1):45-54. doi: 10.1101/gr.3883606. Epub 2005 Dec 12. Genome Res. 2006. PMID: 16344557 Free PMC article. - Trans-natural antisense transcripts including noncoding RNAs in 10 species: implications for expression regulation.
Li JT, Zhang Y, Kong L, Liu QR, Wei L. Li JT, et al. Nucleic Acids Res. 2008 Sep;36(15):4833-44. doi: 10.1093/nar/gkn470. Epub 2008 Jul 24. Nucleic Acids Res. 2008. PMID: 18653530 Free PMC article. - Sense-antisense gene pairs: sequence, transcription, and structure are not conserved between human and mouse.
Wood EJ, Chin-Inmanu K, Jia H, Lipovich L. Wood EJ, et al. Front Genet. 2013 Sep 26;4:183. doi: 10.3389/fgene.2013.00183. eCollection 2013. Front Genet. 2013. PMID: 24133500 Free PMC article. - Economy of Effort or Sophisticated Programming? The Prevalence of Bidirectional Promoter Complexes in the Human Genome.
Anderson EM, Anderson SK. Anderson EM, et al. Genes (Basel). 2024 Feb 18;15(2):252. doi: 10.3390/genes15020252. Genes (Basel). 2024. PMID: 38397241 Free PMC article. Review. - Bidirectional promoters in the transcription of mammalian genomes.
Orekhova AS, Rubtsov PM. Orekhova AS, et al. Biochemistry (Mosc). 2013 Apr;78(4):335-41. doi: 10.1134/S0006297913040020. Biochemistry (Mosc). 2013. PMID: 23590436 Review.
Cited by
- Transcription initiation mapping in 31 bovine tissues reveals complex promoter activity, pervasive transcription, and tissue-specific promoter usage.
Goszczynski DE, Halstead MM, Islas-Trejo AD, Zhou H, Ross PJ. Goszczynski DE, et al. Genome Res. 2021 Apr;31(4):732-744. doi: 10.1101/gr.267336.120. Epub 2021 Mar 15. Genome Res. 2021. PMID: 33722934 Free PMC article. - Activity-dependent human brain coding/noncoding gene regulatory networks.
Lipovich L, Dachet F, Cai J, Bagla S, Balan K, Jia H, Loeb JA. Lipovich L, et al. Genetics. 2012 Nov;192(3):1133-48. doi: 10.1534/genetics.112.145128. Epub 2012 Sep 7. Genetics. 2012. PMID: 22960213 Free PMC article. - Denoising DNA deep sequencing data-high-throughput sequencing errors and their correction.
Laehnemann D, Borkhardt A, McHardy AC. Laehnemann D, et al. Brief Bioinform. 2016 Jan;17(1):154-79. doi: 10.1093/bib/bbv029. Epub 2015 May 29. Brief Bioinform. 2016. PMID: 26026159 Free PMC article. - High-throughput RNA sequencing reveals structural differences of orthologous brain-expressed genes between western lowland gorillas and humans.
Lipovich L, Hou ZC, Jia H, Sinkler C, McGowen M, Sterner KN, Weckle A, Sugalski AB, Pipes L, Gatti DL, Mason CE, Sherwood CC, Hof PR, Kuzawa CW, Grossman LI, Goodman M, Wildman DE. Lipovich L, et al. J Comp Neurol. 2016 Feb 1;524(2):288-308. doi: 10.1002/cne.23843. Epub 2015 Aug 20. J Comp Neurol. 2016. PMID: 26132897 Free PMC article. - A long non-coding RNA GATA6-AS1 adjacent to GATA6 is required for cardiomyocyte differentiation from human pluripotent stem cells.
Jha R, Li D, Wu Q, Ferguson KE, Forghani P, Gibson GC, Xu C. Jha R, et al. FASEB J. 2020 Nov;34(11):14336-14352. doi: 10.1096/fj.202000206R. Epub 2020 Sep 4. FASEB J. 2020. PMID: 32888237 Free PMC article.
References
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. - PubMed
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. - PubMed
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources