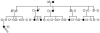Total-genome analysis of BRCA1/2-related invasive carcinomas of the breast identifies tumor stroma as potential landscaper for neoplastic initiation - PubMed (original) (raw)
Total-genome analysis of BRCA1/2-related invasive carcinomas of the breast identifies tumor stroma as potential landscaper for neoplastic initiation
Frank Weber et al. Am J Hum Genet. 2006 Jun.
Abstract
We have shown that the tumor microenvironment of sporadic breast cancer is diverse in genetic alterations and contributes to the cancer phenotype. The dynamic morphology of the mammary gland might be of special interest in hereditary breast/ovarian cancer syndrome (HBOC). We hypothesized that hotspots of loss of heterozygosity or allelic imbalance (LOH/AI) within the tumor stroma of BRCA1/2-related breast cancers provide an impaired mammary stroma that could facilitate later malignant transformation of the breast epithelium. We conducted a total genome LOH/AI scan of DNA derived from the epithelium and stroma of 51 BRCA1/2-related breast cancers, using 372 microsatellite markers. We compared these data with those from a set of 134 sporadic breast cancers. HBOC-related breast cancers accumulated significantly more genetic alterations than did sporadic breast cancers. BRCA1/2-related breast cancer stroma showed LOH/AI at 59.7% of all loci analyzed, similar to the average frequency of LOH/AI observed in the epithelium (66.2%). This is remarkably different from sporadic breast cancers, for which the average epithelial LOH/AI frequency (36.7%) far exceeds the average stromal LOH/AI frequency (28.4%) (P=.03). We identified 11 hotspot loci of LOH/AI in the BRCA1/2 stroma, encompassing genes such as POLD1, which functions in DNA replication, and SDHB. In a subset of samples, enriched for BRCA1 cases, we found 45.0% overall LOH/AI in the stroma, which was significantly higher than the 41.8% LOH/AI observed in corresponding epithelium (P=.04). Together, our data indicate that, in HBOC-related breast cancers, the accumulation of genomic instability in the cancer stroma coincides with that in the neoplastic epithelium, and we postulate that such a genetically unstable stroma might facilitate a microenvironment that functions as a landscaper that promotes genomic instability in the epithelium and, subsequently, neoplastic transformation.
Figures
Figure 1
Pedigree of a family with HBOC segregating BRCA variants. The pedigree is shown across 4 generations, with affected members (with breast cancer) indicated by blackened circles. The proband (arrow) was diagnosed with breast cancer at age 49 years, and testing showed her as wild type for both BRCA genes. Other affected family members (stars) tested positive for the BRCA1 Ser1040Asn and BRCA2 Ser2483Gly variants.
Figure 2
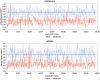
Frequency of LOH/AI observed in the epithelium and stroma in HBOC-related breast cancer compared with sporadic breast cancers. Frequency of LOH/AI (_Y_-axes) is plotted on a markerwise level (_X_-axes). The average LOH/AI frequency of markers in the epithelium (top panel) and stroma (bottom panel) are shown for 51 cases of HBOC-related breast cancer (blue dots) and 134 cases of sporadic breast cancer (red dots). The bold horizontal lines indicate LOH/AI frequencies averaged over all markers.
Figure 3
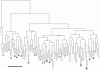
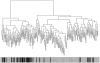
Unsupervised hierarchical cluster analysis. Average linkage and the dissimilarity measure of proportion of discordant LOH/AI between samples are used. The analysis was based on the presence or absence of LOH at 371 informative loci for 51 HBOC-related breast carcinoma epithelial samples and the matching 51 stromal samples (for a total of 102 end branches). The numbers at each end branch indicate germline deleterious mutations in BRCA1 (1) or BRCA2 (2) or unclassified variants of these genes (uv1 and uv2, respectively). The stars indicate those samples for which the stroma and epithelium of one case cluster directly together. Note the clustering of BRCA1 samples near the left (black bar; see text for details). wt = Wild type.
Figure 4
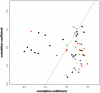
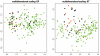
Two-dimensional scaling of 51 HBOC-related breast cancer cases. Each case is represented by combining the stroma and malignant epithelium from the same patient. Samples were obtained from individuals with germline mutations in BRCA1 (black dots) or BRCA2 (red dots), with variants in BRCA1 (BRCA1uv [_green dots_]) or BRCA2 (BRCA2uv [_blue dots_]), or without mutations or variants in either gene (yellow dots). Note the clustering of samples with BRCA1 mutations to the left of the diagonal line.
Figure 5
Multidimensional scaling of LOH/AI patterns from 35 patients with deleterious BRCA1/2 mutations. The LOH/AI patterns from epithelium (EP; left panel) and stroma (ST; right panel) of individuals with germline mutations in BRCA1 (black dots) and BRCA2 (red dots) were analyzed separately in the context of the pattern of LOH/AI observed in sporadic breast cancers (green dots). Note that the stromal LOH/AI pattern differentiates BRCA1 and BRCA2 mutation-positive breast cancers from sporadic breast cancers.
Figure 6
Unsupervised cluster analysis by LOH/AI status in a combined stromal and epithelial data set for HBOC-related breast cancers and sporadic breast cancers. HBOC breast cancers are labeled at the end branches with “DM” (deleterious mutation) and “non-DM” (comprising those with variants of unknown significance and those without detectable mutations). The bar below the cluster plot visualizes the separation of HBOC breast cancers (black and gray bars) from sporadic breast cancers (white bars).
Similar articles
- Novel somatic mutations in the BRCA1 gene in sporadic breast tumors.
Janatova M, Zikan M, Dundr P, Matous B, Pohlreich P. Janatova M, et al. Hum Mutat. 2005 Mar;25(3):319. doi: 10.1002/humu.9308. Hum Mutat. 2005. PMID: 15712267 - Integrated analysis reveals critical genomic regions in prostate tumor microenvironment associated with clinicopathologic phenotypes.
Ashida S, Orloff MS, Bebek G, Zhang L, Zheng P, Peehl DM, Eng C. Ashida S, et al. Clin Cancer Res. 2012 Mar 15;18(6):1578-87. doi: 10.1158/1078-0432.CCR-11-2535. Epub 2012 Jan 24. Clin Cancer Res. 2012. PMID: 22275508 - Combined total genome loss of heterozygosity scan of breast cancer stroma and epithelium reveals multiplicity of stromal targets.
Fukino K, Shen L, Matsumoto S, Morrison CD, Mutter GL, Eng C. Fukino K, et al. Cancer Res. 2004 Oct 15;64(20):7231-6. doi: 10.1158/0008-5472.CAN-04-2866. Cancer Res. 2004. PMID: 15492239 - Involvement of the multiple tumor suppressor genes and 12-lipoxygenase in human prostate cancer. Therapeutic implications.
Gao X, Porter AT, Honn KV. Gao X, et al. Adv Exp Med Biol. 1997;407:41-53. doi: 10.1007/978-1-4899-1813-0_7. Adv Exp Med Biol. 1997. PMID: 9321930 Review. - Clinical management of women with genomic BRCA1 and BRCA2 mutations.
Chang J, Elledge RM. Chang J, et al. Breast Cancer Res Treat. 2001 Sep;69(2):101-13. doi: 10.1023/a:1012203917104. Breast Cancer Res Treat. 2001. PMID: 11759816 Review.
Cited by
- Age-dependent down-regulation of DNA polymerase δ1 in human lymphocytes.
Wang JL, Guo HL, Wang PC, Liu CG. Wang JL, et al. Mol Cell Biochem. 2012 Dec;371(1-2):157-63. doi: 10.1007/s11010-012-1432-6. Epub 2012 Aug 23. Mol Cell Biochem. 2012. PMID: 22915169 - Deregulation of HIF1-alpha and hypoxia-regulated pathways in hepatocellular carcinoma and corresponding non-malignant liver tissue--influence of a modulated host stroma on the prognosis of HCC.
Simon F, Bockhorn M, Praha C, Baba HA, Broelsch CE, Frilling A, Weber F. Simon F, et al. Langenbecks Arch Surg. 2010 Apr;395(4):395-405. doi: 10.1007/s00423-009-0590-9. Epub 2010 Feb 18. Langenbecks Arch Surg. 2010. PMID: 20165955 - BRCA1/2 variants and copy number alterations status in non familial triple negative breast cancer and high grade serous ovarian cancer.
El Ansari FZ, Jouali F, Fekkak R, Bakkach J, Ghailani Nourouti N, Barakat A, Bennani Mechita M, Fekkak J. El Ansari FZ, et al. Hered Cancer Clin Pract. 2022 Aug 19;20(1):29. doi: 10.1186/s13053-022-00236-y. Hered Cancer Clin Pract. 2022. PMID: 35986351 Free PMC article. - Microenvironmental regulation of cancer development.
Hu M, Polyak K. Hu M, et al. Curr Opin Genet Dev. 2008 Feb;18(1):27-34. doi: 10.1016/j.gde.2007.12.006. Epub 2008 Feb 20. Curr Opin Genet Dev. 2008. PMID: 18282701 Free PMC article. Review. - Evaluation of allele-specific somatic changes of genome-wide association study susceptibility alleles in human colorectal cancers.
Gerber MM, Hampel H, Schulz NP, Fernandez S, Wei L, Zhou XP, de la Chapelle A, Ewart Toland A. Gerber MM, et al. PLoS One. 2012;7(5):e37672. doi: 10.1371/journal.pone.0037672. Epub 2012 May 21. PLoS One. 2012. PMID: 22629442 Free PMC article.
References
Web Resources
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for familial breast cancer, BRCA1, BRCA2, SDHB, HIF1α, VEGF, IGF1, POLD1, and IGF2)
- The R Foundation for Statistical Computing, http://www.r-project.org/
References
- American Cancer Society (2005) Breast cancer facts and figures: 2005–2006. American Cancer Society, Atlanta
- Jonsson G, Naylor TL, Vallon-Christersson J, Staaf J, Huang J, Ward MR, Greshock JD, Luts L, Olsson H, Rahman N, Stratton M, Ringner M, Borg A, Weber BL (2005) Distinct genomic profiles in hereditary breast tumors identified by array-based comparative genomic hybridization. Cancer Res 65:7612–7621 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous