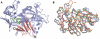Cathepsin D deficiency is associated with a human neurodegenerative disorder - PubMed (original) (raw)
Case Reports
Cathepsin D deficiency is associated with a human neurodegenerative disorder
Robert Steinfeld et al. Am J Hum Genet. 2006 Jun.
Abstract
Cathepsin D is a ubiquitously expressed lysosomal protease that is involved in proteolytic degradation, cell invasion, and apoptosis. In mice and sheep, cathepsin D deficiency is known to cause a fatal neurodegenerative disease. Here, we report a novel disorder in a child with early blindness and progressive psychomotor disability. Two missense mutations in the CTSD gene, F229I and W383C, were identified and were found to cause markedly reduced proteolytic activity and a diminished amount of cathepsin D in patient fibroblasts. Expression of cathepsin D mutants in cathepsin D(-/-) mouse fibroblasts revealed disturbed posttranslational processing and intracellular targeting for W383C and diminished maximal enzyme velocity for F229I. The structural effects of cathepsin D mutants were estimated by computer modeling, which suggested larger structural alterations for W383C than for F229I. Our studies broaden the group of human neurodegenerative disorders and add new insight into the cellular functions of human cathepsin D.
Figures
Figure 1
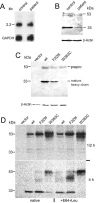
Expression of human mutant and wild-type CatD. A, Northern-blot analysis of CatD mRNA expression. Five micrograms of total RNA purified from control fibroblasts and patient fibroblasts was fractionated by agarose-formaldehyde gel electrophoresis, was blotted, and then was hybridized with a 32P-labeled CatD-specific probe. The amount of CatD-specific mRNA was similar for control and patient RNA, and no additional transcript was detectable in the patient. The quality of mRNA was controlled by equal expression levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). B, Immunodetection of CatD in control and patient fibroblasts. Cell homogenates (50 μg protein per lane) from control and patient human skin fibroblasts were separated by SDS-PAGE, were transferred to nitrocellulose membrane, and were incubated with goat anti-human CatD antibody, then with biotin SP–conjugated donkey anti-goat antibody, and finally with peroxidase-conjugated streptavidin. The blot was stained with TMB substrate to visualize the 53-kDa proenzyme precursor and the 33-kDa heavy chain of mature CatD. The 33-kDa band of patient cells has only 15% of the intensity of band for control cells. The level of β-actin expression is equal for both. C, Expression of human wild-type (wt) CatD and mutants in CatD−/− mouse fibroblasts. Lysates (30 μg protein per lane) from CatD−/− mouse fibroblasts stably transfected with either vector alone or constructs containing human wild-type CatD or mutant F229I and W383C CatD were subjected to SDS-PAGE, were transferred to nitrocellulose membrane, and were stained with TMB substrate after incubation with goat anti-human CatD antibody and peroxidase-conjugated donkey anti-goat antibody. No immunostaining was detectable for vector-transfected cells. All others showed immunoreactivity for the 53-kDa CatD precursor. Wild type–transfected cells had the strongest signal for the 33-kDa heavy chain of mature CatD, mutant F229I showed about 50% of the intensity of the wild type, and no 33-kDa protein was detectable for mutant W383C. The positions of the precursor proenzyme (prepro) and the heavy chain of mature CatD are indicated. D, Metabolic labeling of human wild-type (wt) and mutant CatD. Vector-transfected CatD−/− cells and CatD−/− cells transfected with human wild-type CatD or mutant F229I and W383C CatD were metabolically labeled with [35S]Met for 30 min and were chased for 30 min and 4 h in DMEM supplemented with 10% (v/v) FCS. The CatD was immunoprecipitated from cell lysate using goat anti-human CatD antibody in combination with pansorbin cells and was resolved on SDS-PAGE. Mutant F229I is incompletely processed to the 33-kDa heavy chain of the mature protease when compared with wild-type CatD, whereas mutant W383C is hardly processed at all. After addition of E64 and leupeptin (E64+Leu), the intensity of the 33-kDa heavy chain slightly decreased for all three constructs.
Figure 2
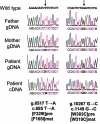
Sequencing data of the two CTSD mutations g.6517T→A and g.10267G→C. By use of genomic DNA templates and either the primer combination hCDgDNA6960F and hCDgDNA7525R or hCDgDNA10788F and hCDgDNA11224R, PCR fragments of the human CTSD gene were amplified and sequenced with both primers independently. The g.6517T→A allele was detected in the mother, the g.10267G→C allele in the father. The patient is compound heterozygous for both alleles. Amplification of CTSD cDNA fragments of the patient (with primers hCDcDNA526F and hCDcDNA1319R) disclosed the presence of the c.685T→A and c.1149G→C alleles in heterozygous form, confirming the results obtained from the sequencing of genomic DNA. The mutations c.685T→A and c.1149G→C result in missense mutations F229I and W383, respectively, of the precursor proenzyme of human CatD (pre) and correspond to amino acid exchanges F165I and W319C, respectively, of the mature human CatD (mat).
Figure 3
Alignment of the mature human CatD with selected active peptidases of the pepsin family. The amino acid sequence of human CatD (h-cathepsin D [aa 66–410]) was aligned with the following pepsin peptidases: human pepsin A and human renin (h-pepsin A [aa 63–388] and h-renin [aa 73–406]), saccharopepsin (aa 78–405) from the yeast Saccharomyces cerevisiae, mucorpepsin (aa 76–424) from the fungus Rhizomucor miehei, oryzepsin (aa 72–390) from the fungus Aspergillus oryzae, plasmepsin-1 (aa 126–449) from the sporozoan Plasmodium falciparum, and CDR1 g.p. (aa 70–437) from the plant Arabidopsis thaliana. The sequences were retrieved from the MEROPS database and were ordered by degree of homology. Identical amino acid residues are red, highly homologous amino acids are green, and low-homology amino acids are blue. A characteristic feature of the pepsin family is the three amino acids D39, Y84, and D261 (asterisks [*]). In the h-cathepsin D sequence, the 8-aa residues that separate the N-terminal light chain (97 aa) from the C-terminal heavy chain (244 aa) are underlined. During the maturation of human CatD, these 8 aa are cleaved off. The positions of the two human missense mutations found in the patient are indicated by arrows. F183 (F229 in the full-length h-cathepsin D sequence) is strictly conserved, W358 (W383 in the full-length h-cathepsin D sequence) is merely conserved among human peptidases of the pepsin family (shown only for h-pepsin A and h-renin).
Figure 4
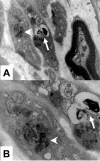
Electron microscopy of storage material in Schwann cells from skin biopsy specimen from the patient. A, At a magnification of 20,000×, the image displays two nonmyelinated Schwann cells (left) and one myelinated Schwann cell. The two nonmyelinated Schwann cells contain various osmiophilic inclusions (arrow and arrowhead). B, Displayed details at 50,000× magnification show two types of intracellular inclusions: granular osmiophilic deposits (arrowhead) and myelin-like lamellar structures (arrow).
Figure 5
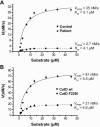
Kinetic analysis of human CatD in patient fibroblasts and transfected CatD−/− mouse fibroblasts. The kinetic properties of human CatD were measured for patient and control lysates (A) and for lysates derived from CatD−/− mouse fibroblasts transfected with wild-type CatD (wt) and mutant F229I (B). Increasing concentrations of the fluorogenic substrate MOCAc-GKPIIFFRLK(Dnp)-R-NH2 were incubated for 20 min with 0.5 μg of cell lysates each. The increase in fluorescence was plotted in relation to the concentration of substrate, and _K_m and _V_max were determined by nonlinear curve fitting. The calculated _K_m values were similar to the reported value (
K _m_=3.7
μM) determined with purified CatD. _V_max for patient lysates was only 7.7% of that measured for control lysates. Lysates derived from CatD−/− mouse fibroblasts transfected with mutant F229I had a _V_max of 26% when compared with those transfected with wild-type CatD.
Figure 6
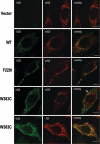
Immunohistochemical localization of CatD mutants expressed in Flp-In 3T3 cells and CatD−/− mouse fibroblasts. The cells were cultivated on glass coverslips. Washed and fixed cells were costained with goat anti-human CatD antibody and secondary Alexa Fluor 488 conjugates (green) and with either rabbit anti-mouse CatD antibody or with mouse anti–protein disulfide isomerase (PDI) antibody and secondary Alexa Fluor 546 conjugates (red). Vector, No signal for human CatD (hCD); only mouse CatD (mCD) is detectable with typical pattern (red). WT, In 3T3 cells transfected with human wild-type CatD, human and mouse CatD colocalize. F229I, In 3T3 cells transfected with mutant F229I CatD, human and mouse CatD colocalize. W383C, In 3T3 cells transfected with mutant W383C, the staining pattern for human CatD (green) clearly differs from the signal for mouse CatD (red). However, minor signals show colocalization of mutant W383C with mouse CatD (arrow). W383C, In CatD−/− cells transfected with mutant W383C, the staining pattern for human CatD (green) mostly colocalizes with the ER marker (red; anti-PDI antibody). The length of the bars corresponds to 8 μm.
Figure 7
Structural effects of CTSD mutations. A, Cartoon structure of CatD showing the positions of mutations within the enzyme domains. The active site is painted as a blue transparent surface. N and C labels indicate the N and C termini. The interdomain β-sheet of CatD and its flanking regions are in pink. This interdomain β-sheet plays a crucial role in the regulation of the enzymatic activity of CatD. Its N-terminal strand (numbered 1) was shown to relocate at neutral pH and to insert into the active site cleft, effectively blocking substrate access and keeping CatD in an inactive state. The residue F165 (F [_red_]) is located within strand 3 of the interdomain β-sheet. The N-terminal region of strand 4 is flanked by a short α-helix (aa G322 to Y329 [_light green_]) and by W319 (W [_green_]). B, Least-squares fit for the three averaged structures of wild-type CatD (blue), F165I (green), and W319C (red). N and C labels indicate the N and C termini. There are no major structural differences between the backbone coordinates of wild-type CatD and the F165I mutant (average root mean SD 1.31 Å). The comparison of main-chain atom coordinates of wild-type CatD and those of the W319C mutant revealed higher differences (average root mean SD 2.02 Å), with a maximum deviation of ∼10.1 Å for G298 (red arrow). Note that amino acid residues are assigned on the basis of the published crystal structure, and these residues correspond to residues of the full-length CatD precursor proenzyme sequence as follows: F165I = F229I, G298 = G362, W319C = W383C, G322 = G386, and Y329 = Y393.
Similar articles
- Enzyme replacement therapy with recombinant pro-CTSD (cathepsin D) corrects defective proteolysis and autophagy in neuronal ceroid lipofuscinosis.
Marques ARA, Di Spiezio A, Thießen N, Schmidt L, Grötzinger J, Lüllmann-Rauch R, Damme M, Storck SE, Pietrzik CU, Fogh J, Bär J, Mikhaylova M, Glatzel M, Bassal M, Bartsch U, Saftig P. Marques ARA, et al. Autophagy. 2020 May;16(5):811-825. doi: 10.1080/15548627.2019.1637200. Epub 2019 Jul 16. Autophagy. 2020. PMID: 31282275 Free PMC article. - Progranulin-mediated deficiency of cathepsin D results in FTD and NCL-like phenotypes in neurons derived from FTD patients.
Valdez C, Wong YC, Schwake M, Bu G, Wszolek ZK, Krainc D. Valdez C, et al. Hum Mol Genet. 2017 Dec 15;26(24):4861-4872. doi: 10.1093/hmg/ddx364. Hum Mol Genet. 2017. PMID: 29036611 Free PMC article. - The Role of Cathepsin D in the Pathogenesis of Human Neurodegenerative Disorders.
Vidoni C, Follo C, Savino M, Melone MA, Isidoro C. Vidoni C, et al. Med Res Rev. 2016 Sep;36(5):845-70. doi: 10.1002/med.21394. Epub 2016 Apr 26. Med Res Rev. 2016. PMID: 27114232 Review. - Recombinant pro-CTSD (cathepsin D) enhances SNCA/α-Synuclein degradation in α-Synucleinopathy models.
Prieto Huarcaya S, Drobny A, Marques ARA, Di Spiezio A, Dobert JP, Balta D, Werner C, Rizo T, Gallwitz L, Bub S, Stojkovska I, Belur NR, Fogh J, Mazzulli JR, Xiang W, Fulzele A, Dejung M, Sauer M, Winner B, Rose-John S, Arnold P, Saftig P, Zunke F. Prieto Huarcaya S, et al. Autophagy. 2022 May;18(5):1127-1151. doi: 10.1080/15548627.2022.2045534. Epub 2022 Apr 28. Autophagy. 2022. PMID: 35287553 Free PMC article. - Proteolytic activation of human procathepsin D.
Richo G, Conner GE. Richo G, et al. Adv Exp Med Biol. 1991;306:289-96. doi: 10.1007/978-1-4684-6012-4_35. Adv Exp Med Biol. 1991. PMID: 1812719 Review.
Cited by
- Whole exome screening of neurodevelopmental regression disorders in a cohort of Egyptian patients.
Refeat MM, Naggar WE, Saied MME, Kilany A. Refeat MM, et al. Neurogenetics. 2023 Jan;24(1):17-28. doi: 10.1007/s10048-022-00703-7. Epub 2022 Nov 26. Neurogenetics. 2023. PMID: 36435927 Free PMC article. - Prospects of β-Secretase Inhibitors for the Treatment of Alzheimer's Disease.
Ghosh AK, Tang J. Ghosh AK, et al. ChemMedChem. 2015 Sep;10(9):1463-6. doi: 10.1002/cmdc.201500216. Epub 2015 Jul 3. ChemMedChem. 2015. PMID: 26140607 Free PMC article. - Haematopoietic development and immunological function in the absence of cathepsin D.
Tulone C, Uchiyama Y, Novelli M, Grosvenor N, Saftig P, Chain BM. Tulone C, et al. BMC Immunol. 2007 Sep 26;8:22. doi: 10.1186/1471-2172-8-22. BMC Immunol. 2007. PMID: 17897442 Free PMC article. - CLN7 is an organellar chloride channel regulating lysosomal function.
Wang Y, Zeng W, Lin B, Yao Y, Li C, Hu W, Wu H, Huang J, Zhang M, Xue T, Ren D, Qu L, Cang C. Wang Y, et al. Sci Adv. 2021 Dec 17;7(51):eabj9608. doi: 10.1126/sciadv.abj9608. Epub 2021 Dec 15. Sci Adv. 2021. PMID: 34910516 Free PMC article. - Cathepsin D in Podocytes Is Important in the Pathogenesis of Proteinuria and CKD.
Yamamoto-Nonaka K, Koike M, Asanuma K, Takagi M, Oliva Trejo JA, Seki T, Hidaka T, Ichimura K, Sakai T, Tada N, Ueno T, Uchiyama Y, Tomino Y. Yamamoto-Nonaka K, et al. J Am Soc Nephrol. 2016 Sep;27(9):2685-700. doi: 10.1681/ASN.2015040366. Epub 2016 Jan 28. J Am Soc Nephrol. 2016. PMID: 26823550 Free PMC article.
References
Web Resources
- MEROPS—the Peptidase Database, http://merops.sanger.ac.uk/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CONCL) - PubMed
- Protein Data Bank, http://www.rcsb.org/pdb/
References
- Liaudet-Coopman E, Beaujouin M, Derocq D, Garcia M, Glondu-Lassis M, Laurent-Matha V, Prebois C, Rochefort H, Vignon F (2005) Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett (http://www.sciencedirect.com/science/journal/03043835) (electronically published July 18, 2005; accessed February 24, 2006) - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous