Batai and Ngari viruses: M segment reassortment and association with severe febrile disease outbreaks in East Africa - PubMed (original) (raw)
Batai and Ngari viruses: M segment reassortment and association with severe febrile disease outbreaks in East Africa
Thomas Briese et al. J Virol. 2006 Jun.
Abstract
Ngari virus is an orthobunyavirus recently recognized as a reassortant between Bunyamwera virus and an as yet unidentified M segment donor. Analysis of M segment sequences of Batai and Ilesha viruses revealed 95% deduced amino acid identity between Batai virus and Ngari virus. These findings suggest Batai virus as the donor of Ngari virus M segment sequence. Analysis of Batai virus-related African isolates identified UgMP-6830, isolated from mosquitoes in Uganda, as an isolate of Batai virus. KV-141, isolated during a febrile disease outbreak in Sudan, was identified as another isolate of Ngari virus, emphasizing a role of this reassortant virus in severe human illness throughout East Africa.
Figures
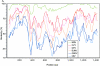
FIG.1.
Analysis of M segment sequence conservation between BATV, NRIV, and ILEV. (A) Sliding-window distance analysis between BATV (MM2222) and NRIV (Kenya), ILEV, CVV, BUNV, Guaroa virus (GROV; GenBank accession no. AY380581), and California encephalitis virus (CEV; AF123483) (window, 60 amino acids [aa]; step 10 amino acids). (B) Alignment of terminal M segment sequences of BATV (MM2222), NRIV (Kenya), and ILEV. Coding sequence for the polyprotein is omitted, and only the ochre codon and initiation codon are indicated (underlined and overlined UUA and CAU, respectively; sequence given in genomic antisense orientation). Sequences used in PCR priming are indicated in lowercase italics.
FIG.1.
Analysis of M segment sequence conservation between BATV, NRIV, and ILEV. (A) Sliding-window distance analysis between BATV (MM2222) and NRIV (Kenya), ILEV, CVV, BUNV, Guaroa virus (GROV; GenBank accession no. AY380581), and California encephalitis virus (CEV; AF123483) (window, 60 amino acids [aa]; step 10 amino acids). (B) Alignment of terminal M segment sequences of BATV (MM2222), NRIV (Kenya), and ILEV. Coding sequence for the polyprotein is omitted, and only the ochre codon and initiation codon are indicated (underlined and overlined UUA and CAU, respectively; sequence given in genomic antisense orientation). Sequences used in PCR priming are indicated in lowercase italics.
Similar articles
- Ngari virus is a Bunyamwera virus reassortant that can be associated with large outbreaks of hemorrhagic fever in Africa.
Gerrard SR, Li L, Barrett AD, Nichol ST. Gerrard SR, et al. J Virol. 2004 Aug;78(16):8922-6. doi: 10.1128/JVI.78.16.8922-8926.2004. J Virol. 2004. PMID: 15280501 Free PMC article. - Genetic characterization of Batai virus indicates a genomic reassortment between orthobunyaviruses in nature.
Yanase T, Kato T, Yamakawa M, Takayoshi K, Nakamura K, Kokuba T, Tsuda T. Yanase T, et al. Arch Virol. 2006 Nov;151(11):2253-60. doi: 10.1007/s00705-006-0808-x. Epub 2006 Jul 6. Arch Virol. 2006. PMID: 16820982 - Mammals Preferred: Reassortment of Batai and Bunyamwera orthobunyavirus Occurs in Mammalian but Not Insect Cells.
Heitmann A, Gusmag F, Rathjens MG, Maurer M, Frankze K, Schicht S, Jansen S, Schmidt-Chanasit J, Jung K, Becker SC. Heitmann A, et al. Viruses. 2021 Aug 27;13(9):1702. doi: 10.3390/v13091702. Viruses. 2021. PMID: 34578285 Free PMC article. - [ZOOPATHOGENIC ORTHOBUNIAVIRUSES (ORTHOBUNYAVIRUS, BUNYAVIRIDAE)].
Makarov VV, Guliukin MI, Lvov DK. Makarov VV, et al. Vopr Virusol. 2016;61(2):53-8. Vopr Virusol. 2016. PMID: 27451495 Review. Russian. - A Review of Bunyamwera, Batai, and Ngari Viruses: Understudied Orthobunyaviruses With Potential One Health Implications.
Dutuze MF, Nzayirambaho M, Mores CN, Christofferson RC. Dutuze MF, et al. Front Vet Sci. 2018 Apr 12;5:69. doi: 10.3389/fvets.2018.00069. eCollection 2018. Front Vet Sci. 2018. PMID: 29707545 Free PMC article. Review.
Cited by
- Monitoring for bovine arboviruses in the most southwestern islands in Japan between 1994 and 2014.
Kato T, Yanase T, Suzuki M, Katagiri Y, Ikemiyagi K, Takayoshi K, Shirafuji H, Ohashi S, Yoshida K, Yamakawa M, Tsuda T. Kato T, et al. BMC Vet Res. 2016 Jun 24;12(1):125. doi: 10.1186/s12917-016-0747-z. BMC Vet Res. 2016. PMID: 27342576 Free PMC article. - VIPR HMM: a hidden Markov model for detecting recombination with microbial detection microarrays.
Allred AF, Renshaw H, Weaver S, Tesh RB, Wang D. Allred AF, et al. Bioinformatics. 2012 Nov 15;28(22):2922-9. doi: 10.1093/bioinformatics/bts560. Epub 2012 Oct 7. Bioinformatics. 2012. PMID: 23044542 Free PMC article. - Granada virus: a natural phlebovirus reassortant of the sandfly fever Naples serocomplex with low seroprevalence in humans.
Collao X, Palacios G, de Ory F, Sanbonmatsu S, Pérez-Ruiz M, Navarro JM, Molina R, Hutchison SK, Lipkin WI, Tenorio A, Sánchez-Seco MP. Collao X, et al. Am J Trop Med Hyg. 2010 Oct;83(4):760-5. doi: 10.4269/ajtmh.2010.09-0697. Am J Trop Med Hyg. 2010. PMID: 20889862 Free PMC article. - It's in the mix: Reassortment of segmented viral genomes.
Lowen AC. Lowen AC. PLoS Pathog. 2018 Sep 13;14(9):e1007200. doi: 10.1371/journal.ppat.1007200. eCollection 2018 Sep. PLoS Pathog. 2018. PMID: 30212586 Free PMC article. Review. No abstract available. - Evidence of circulation of Orthobunyaviruses in diverse mosquito species in Kwale County, Kenya.
Koka H, Lutomiah J, Langat S, Koskei E, Nyunja A, Mutisya J, Mulwa F, Owaka S, Ofula V, Konongoi S, Eyase F, Sang R. Koka H, et al. Virol J. 2021 Oct 12;18(1):204. doi: 10.1186/s12985-021-01670-5. Virol J. 2021. PMID: 34641884 Free PMC article.
References
- Bardos, V., and E. Cupkova. 1962. The Calovo virus—the second virus isolated from mosquitoes in Czechoslovakia. J. Hyg. Epidemiol. Microbiol. Immunol. 6:186-192. - PubMed
- Beaty, B. J., D. R. Sundin, L. J. Chandler, and D. H. Bishop. 1985. Evolution of bunyaviruses by genome reassortment in dually infected mosquitoes (Aedes triseriatus). Science 230:548-550. - PubMed
- Borucki, M. K., L. J. Chandler, B. M. Parker, C. D. Blair, and B. J. Beaty. 1999. Bunyavirus superinfection and segment reassortment in transovarially infected mosquitoes. J. Gen. Virol. 80:3173-3179. - PubMed
- Bowen, M. D., S. G. Trappier, A. J. Sanchez, R. F. Meyer, C. S. Goldsmith, S. R. Zaki, L. M. Dunster, C. J. Peters, T. G. Ksiazek, and S. T. Nichol. 2001. A reassortant bunyavirus isolated from acute hemorrhagic fever cases in Kenya and Somalia. Virology 291:185-190. - PubMed
- Briese, T., A. Rambaut, and W. I. Lipkin. 2004. Analysis of the medium (M) segment sequence of Guaroa virus and its comparison to other orthobunyaviruses. J. Gen. Virol. 85:3071-3077. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources