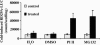The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1 - PubMed (original) (raw)
The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1
Chun-Hai Dong et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Plant responses to cold stress are mediated by a transcriptional cascade, in which the transcription factor ICE1 and possibly related proteins activate the expression of C-repeat (CRT)-binding factors (CBFs), leading to the transcription of downstream effector genes. The variant RING finger protein high expression of osmotically responsive gene (HOS)1 was identified genetically as a negative regulator of cold responses. We present evidence here that HOS1 is an E3 ligase required for the ubiquitination of ICE1. HOS1 physically interacts with ICE1 and mediates the ubiquitination of ICE1 both in vitro and in vivo. We found that cold induces the degradation of ICE1 in plants, and this degradation requires HOS1. Consistent with enhanced cold-responsive gene expression in loss-of-function hos1 mutant plants, overexpression of HOS1 represses the expression of CBFs and their downstream genes and confers increased sensitivity to freezing stress. Our results indicate that cold stress responses in Arabidopsis are attenuated by a ubiquitination/proteasome pathway in which HOS1 mediates the degradation of the ICE1 protein.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Fig. 1.
Proteasome inhibitors enhance cold-induced expression of RD29A-LUC. Arabidopsis seeds (wild type containing RD29A-LUC) were germinated and grown on Murashige and Skoog plates for 10 days. Half of the seedlings were pretreated for 24 h in a Petri dish with H2O, 2% DMSO, or 40 μM proteasome inhibitors (PI II and MG132, Calbiochem) in 2% DMSO (treated). The untreated seedlings were used as controls. Luminescence was analyzed after cold treatment at 0°C for 24 h. Shown are the mean values + SE from 20 individual seedlings.
Fig. 2.
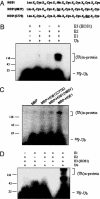
E3 activity of HOS1. (A) Amino acid sequence comparison of HOS1, HOS1(H63Y), and HOS1(C77S) in the RING motif. The mutated residues in HOS1(H63Y) and HOS1(C77S) are underlined. (B) MBP-HOS1 was assayed for E3 activity in the presence of E1, E2, and 32P-labeled ubiquitin (Ub), as indicated. Radiolabeled bands corresponding to free ubiquitin (32P-Ub) and polyubiquitinated MBP-HOS1 ((Ub)n-HOS1) are indicated. (C) MBP-HOS1 E3 activity depends on its RING motif. Wild-type MBP-HOS1 and MBP-HOS1 mutants (H63Y and C77S) were assayed for self-ubiquitination. (D) Ubiquitination assay of HOS1 E3 ligase at 4°C in the presence of E1, E2, and 32P-labeled ubiquitin (Ub), as indicated.
Fig. 3.
The RING domain of HOS1 is essential but not sufficient for its E3 ligase activity. (A) Diagram of the HOS1 proteins used for the activity assay. The RING domain is indicated in wild-type HOS1 and truncated HOS1 mutants. (B) Self-ubiquitination assay of wild-type MBP-HOS1 and MBP-HOS1 truncated mutants (1–210, 1–550). MBP is included as a negative control. (C) E3 ligase activity of HOS1 and CBL. The reaction temperatures (30°C or 4°C) are indicated.
Fig. 4.
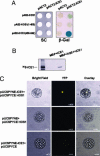
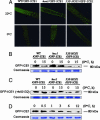
RING finger protein HOS1 interacts with ICE1. (A) HOS1 interacts with ICE1 in the yeast two-hybrid assay. Wild-type HOS1 interacts with ICE1 weakly, but the HOS1 C-terminal region (amino acids 450–842), not the N-terminal region (amino acids 1–450), interacts with ICE1 strongly. Yeast strains containing the pAS2-HOS1, pAS2-HOS1 (1–450), and pAS2-HOS1 (450–842) bait and the pACT2 or pACT2-ICE1 prey were assayed for β-gal activity (blue color). The pAS2-HOS1/pACT2, pAS2-HOS1(1–450)/pACT2, and pAS2-HOS1(450–842)/pACT2 combinations were used as negative controls. Yeast grown on synthetic complete (SC) plate (Left) and the corresponding β-gal filter assay (Right) are shown. (B) HOS1 binds to ICE1 in vitro. Recombinant MBP-HOS1 but not MBP could pull down _in vitro_-translated 35S-ICE1. The first lane from the left shows _in vitro_-translated 35S-ICE1. (C) HOS1 interacts with ICE1 in vivo. pUCSPYNE-ICE1 was transiently coexpressed with pUCSPYCE-HOS1 or the pUCSPYCE vector, and pUCSPYCE-HOS1 was transiently coexpressed with the pUCSPYNE vector in wild-type Arabidopsis protoplasts. YFP fluorescence was detected only when pUCSPYNE-ICE1 was coexpressed with pUCSPYCE-HOS1.
Fig. 5.
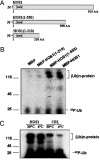
Ubiquitination of ICE1 in vitro and in vivo. (A) Ubiquitination of ICE1 by HOS1 in vitro. 35S-ICE1 was assayed for in vitro ubiquitination in the presence of E1, E2, and E3 (MBP-HOS1). Polyubiquitinated ICE1 is indicated as (Ub)n-ICE1. (B) Detection of in vivo polyubiquitinated GFP-ICE1 in wild-type and hos1 plants after cold treatment. Seedlings of GFP-ICE1 in wild-type (WT/GFP-ICE1) or hos1 mutant (hos1/GFP-ICE1) background were pretreated with the proteasome inhibitor MG132 (50 μM) at 4°C for 15 h. Total protein extract from 30 seedlings was immunoprecipitated with antibody against GFP and analyzed by immunoblotting with antibody against ubiquitin. A Coomassie blue-stained nonspecific band from a duplicate gel is shown as loading control.
Fig. 6.
Cold-induced degradation of GFP-ICE1. (A) Visualization of GFP-ICE1 fusion protein. Ten-day-old seedlings of wild-type (WT/GFP-ICE1), hos1 (hos1/GFP-ICE1), and 35S-HOS1 (35S-HOS1/GFP-ICE1) plants grown on agar plates were treated (0°C, 15 h) or not treated (22°C) with cold stress. Seedling roots were observed immediately after cold stress under a confocal microscope. (B) Detection of GFP-ICE1 protein by Western blot analysis. Wild-type (WT/GFP-ICE1), hos1 (hos1/GFP-ICE1), or 35S-HOS1 (35S-HOS1/GFP-ICE1) seedlings were treated with cold, as described above. GFP-ICE1 protein (80-kDa) levels were analyzed with anti-GFP. (C) Detection of GFP-ICE1 protein levels in the presence of MG132. Wild-type (WT/GFP-ICE1), hos1 (hos1/GFP-ICE1), or 35S-HOS1 (35S-HOS1/GFP-ICE1) seedlings were treated with cold in the presence of MG132 (50 μM). GFP-ICE1 protein (80 kDa) was analyzed with anti-GFP. (D) Kinetics of GFP-ICE1 degradation upon cold treatment. Ten-day-old wild-type (WT/GFP-ICE1) seedlings were treated at 0°C for different times (0, 0.5, 1, 3, 6, or 12 h), as indicated. Levels of the GFP-ICE1 protein (80 kDa) were analyzed with anti-GFP. In B–D, a nonspecific Coomassie blue-stained band from a duplicate gel shows that similar amounts of proteins were loaded.
Fig. 7.
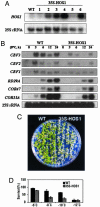
Gene expression analysis and freezing sensitivity of _HOS1-_overexpression plants. (A) Overexpression of HOS1 in transgenic Arabidopsis plants. Total RNA was prepared from wild-type (WT) and 35S-HOS1 transgenic seedlings. The blot was probed with labeled HOS1 cDNA or 25S rRNA as a control. (B) Transcript levels of CBF and the CBF downstream target genes in WT and HOS1-overexpression plants (35S-HOS1). Gene expression was analyzed by Northern blot hybridization with total RNA prepared from 2-week-old WT and HOS1-overexpression transgenic Arabidopsis plants after cold treatment. The blot was probed with labeled CBF3, CBF2, CBF1, RD29A, COR47, COR15A, and 25S rRNA, respectively. (C) Decreased survival of _HOS1-_overexpression transgenic plants (35S-HOS1) after a freezing treatment at −8°C. (D) Quantification of the survival rates for WT and HOS1-ovexpression transgenic plants (35S-HOS1) upon freezing treatment, after 48-h cold acclimation at 4°C. Data shown are mean values with standard errors (n = 7). The two genotypes are significantly different (χ2 test, P < 0.01) at all of the tested temperatures.
Similar articles
- The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis.
Kim YS, Lee M, Lee JH, Lee HJ, Park CM. Kim YS, et al. Plant Mol Biol. 2015 Sep;89(1-2):187-201. doi: 10.1007/s11103-015-0365-3. Epub 2015 Aug 27. Plant Mol Biol. 2015. PMID: 26311645 - OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis.
Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S. Ding Y, et al. Dev Cell. 2015 Feb 9;32(3):278-89. doi: 10.1016/j.devcel.2014.12.023. Dev Cell. 2015. PMID: 25669882 - ICE1 Ser403 is necessary for protein stabilization and regulation of cold signaling and tolerance.
Miura K, Ohta M, Nakazawa M, Ono M, Hasegawa PM. Miura K, et al. Plant J. 2011 Jul;67(2):269-79. doi: 10.1111/j.1365-313X.2011.04589.x. Epub 2011 May 9. Plant J. 2011. PMID: 21447070 - Molecular genetic analysis of cold-regulated gene transcription.
Viswanathan C, Zhu JK. Viswanathan C, et al. Philos Trans R Soc Lond B Biol Sci. 2002 Jul 29;357(1423):877-86. doi: 10.1098/rstb.2002.1076. Philos Trans R Soc Lond B Biol Sci. 2002. PMID: 12171651 Free PMC article. Review. - CBF-dependent signaling pathway: a key responder to low temperature stress in plants.
Zhou MQ, Shen C, Wu LH, Tang KX, Lin J. Zhou MQ, et al. Crit Rev Biotechnol. 2011 Jun;31(2):186-92. doi: 10.3109/07388551.2010.505910. Epub 2010 Oct 4. Crit Rev Biotechnol. 2011. PMID: 20919819 Review.
Cited by
- Genomic identification and expression analysis of nuclear pore proteins in Malus domestica.
Zhang C, An N, Jia P, Zhang W, Liang J, Zhang X, Zhou H, Ma W, Han M, Xing L, Ren X. Zhang C, et al. Sci Rep. 2020 Oct 15;10(1):17426. doi: 10.1038/s41598-020-74171-0. Sci Rep. 2020. PMID: 33060661 Free PMC article. - ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco.
Wu L, Chen X, Ren H, Zhang Z, Zhang H, Wang J, Wang XC, Huang R. Wu L, et al. Planta. 2007 Sep;226(4):815-25. doi: 10.1007/s00425-007-0528-9. Epub 2007 May 4. Planta. 2007. PMID: 17479283 - Identification of multiple novel genetic mechanisms that regulate chilling tolerance in Arabidopsis.
Sahoo DK, Hegde C, Bhattacharyya MK. Sahoo DK, et al. Front Plant Sci. 2023 Jan 12;13:1094462. doi: 10.3389/fpls.2022.1094462. eCollection 2022. Front Plant Sci. 2023. PMID: 36714785 Free PMC article. - Transcriptome analysis of the winter wheat Dn1 in response to cold stress.
Tian Y, Peng K, Lou G, Ren Z, Sun X, Wang Z, Xing J, Song C, Cang J. Tian Y, et al. BMC Plant Biol. 2022 Jun 6;22(1):277. doi: 10.1186/s12870-022-03654-1. BMC Plant Biol. 2022. PMID: 35659183 Free PMC article. - Integration of low temperature and light signaling during cold acclimation response in Arabidopsis.
Catalá R, Medina J, Salinas J. Catalá R, et al. Proc Natl Acad Sci U S A. 2011 Sep 27;108(39):16475-80. doi: 10.1073/pnas.1107161108. Epub 2011 Sep 19. Proc Natl Acad Sci U S A. 2011. PMID: 21930922 Free PMC article.
References
- Thomashow M. F. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:571–599. - PubMed
- Gilmour S. J., Zarka D. G., Stockinger E. J., Salazar M. P., Houghton J. M., Thomashow M. F. Plant J. 1998;16:433–442. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials