ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts - PubMed (original) (raw)
ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts
Ken-ichi Yoshioka et al. Mol Cell. 2006.
Abstract
S(N)1-type alkylating agents that produce cytotoxic O(6)-methyl-G (O(6)-meG) DNA adducts induce cell cycle arrest and apoptosis in a manner requiring the DNA mismatch repair (MMR) proteins MutSalpha and MutLalpha. Here, we show that checkpoint signaling in response to DNA methylation occurs during S phase and requires DNA replication that gives rise to O(6)-meG/T mispairs. DNA binding studies reveal that MutSalpha specifically recognizes O(6)-meG/T mispairs, but not O(6)-meG/C. In an in vitro assay, ATR-ATRIP, but not RPA, is preferentially recruited to O(6)-meG/T mismatches in a MutSalpha- and MutLalpha-dependent manner. Furthermore, ATR kinase is activated to phosphorylate Chk1 in the presence of O(6)-meG/T mispairs and MMR proteins. These results suggest that MMR proteins can act as direct sensors of methylation damage and help recruit ATR-ATRIP to sites of cytotoxic O(6)-meG adducts to initiate ATR checkpoint signaling.
Figures
Figure 1
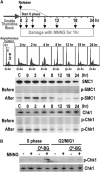
MNNG Damage Signaling Coincides with S Phase (A) Cell cycle dependence of MNNG damage signaling. Synchronized HeLaS3 cells were released from a double thymidine block and cultured for varying times in complete medium followed by incubation for 1 hr in serum-free medium in the presence or absence of 10 μM MNNG prior to harvesting. Synchronization was monitored by FACS prior to MNNG treatment. Phosphorylated and nonphosphorylated SMC1 and Chk1 were detected by immunoblotting. “C” is an asynchronous cell population. (B) DNA damage signaling in response to O6-meG adducts. HeLaS3 cells synchronized in S phase (2 hr after release from a double thymidine block) or G2-M-G1 phases (12 hr after release) were incubated in the presence or absence of O6-benzyl-guanine followed by incubation for 2 hr in serum-free medium containing 2 μM MNNG. Lysates were analyzed by immunoblotting.
Figure 2
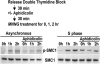
DNA Damage Signaling Triggered by MNNG Modification Depends on DNA Replication HeLaS3 cells, from either unsynchronized or synchronized cultures, were incubated in the presence or absence of aphidicolin followed by the addition of 10 μM MNNG for 1 or 2 hr in serum-free medium. Lysates were analyzed by immunoblotting to monitor phosphorylation of SMC1.
Figure 3
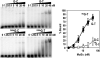
MutSα Binds to O6-meG/T Mispairs Gel mobility shift assays were performed with 41 bp DNA duplexes containing G/C, G/T, O6-meG/C, or O6-meG/T base pairs. 32P 5′ labeled DNA was incubated with the indicated concentration of purified recombinant MutSα and subjected to native gel electrophoresis. Concentrations of MutSα that gave half-maximal binding were 10 ± 0.9 nM for O6-meG/T, 10 ± 1.1 nM for G/T, and >100 nM for O6-meG/C and G/C. Error bars represent the SDs of three independent experiments.
Figure 4
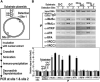
MutSα, MutLα, and ATR-ATRIP Localize to Regions of O6-meG/T Mispairs (A) Scheme for CrIP assay. See the Experimental Procedures for details. The preferred association of MutSα, MutLα, and ATR-ATRIP with various DNA mispairs was determined by the relative recovery of sites 1 and 2 after PCR. (B) MutSα, MutLα, and ATR-ATRIP associate preferentially with an O6-meG/T mispair. Relative recoveries of MutSα, MutLα, ATR/ATRIP, RPA, XRCC2, and XRCC3 at sites 1 and 2 were determined by the CrIP assay for G/C, G/T, O6-meG/C, and O6-meG/T substrates using HeLaS3 nuclear extracts and indicated antisera. Error bars represent the SDs of three independent experiments.
Figure 5
Association of ATR-ATRIP with O6-meG/T Requires Both MutSα and MutLα Recovery of ATR and ATRIP at O6-meG/T-containing DNA was determined in the CrIP assay by using LoVo or HCT116 nuclear extracts either alone or supplemented with purified, recombinant murine MutSα or MutLα as indicated. Error bars represent the SDs of three independent experiments.
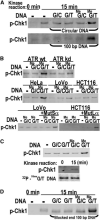
Figure 6
ATR Kinase Activation by O6-meG/T in the Presence of MutSα and MutLα (A) His6-Chk1 phosphorylation dependent on O6-meG/T. Kinase assays with 20 nM recombinant Chk1 and HeLaS3 nuclear extract were carried out with closed circular DNAs or 100 bp duplexes containing G/C, G/T, O6-meG/C, or O6-meG/T base pairs followed by Western blotting with anti-phosphoChk1 antibody. (B) O6-meG/T-dependent phosphorylation of Chk1 requires ATR, MutSα, and MutLα. Kinase assays were determined as in (A) in the absence or presence of 100 bp O6-meG/C or O6-meG/T DNA by using nuclear extracts from U2OS conditionally overexpressing wild-type (wt) or kinase-deficient (kd) ATR, or HeLaS3, LoVo, or HCT116 cells. Where indicated, LoVo or HCT116 nuclear extracts were supplemented with recombinant murine MutSα or MutLα. (C) Recovery of duplex DNA after ATR kinase assay. Top, Chk1 phosphorylation was assayed by using biotinylated 100 bp duplexes containing G/C, G/T, O6-meG/C, or O6-meG/T incubated with HeLaS3 nuclear extracts followed by a pull-down with streptavidin beads. Bottom, biotinylated 100 bp duplexes containing an O6-meG/T mispair and 32P 5′ labeled on both strands were used in a kinase assay after pull-down as described above. 32P 5′ labeled duplex DNA from kinase assays (15 min) or control samples (0 min) was also analyzed by denaturing PAGE. (D) O6-meG/T-dependent His6-Chk1 phosphorylation with blocked end DNA. Kinase assays contained streptavidin-dual-blocked-end-100 bp duplexes containing G/C, G/T, O6-meG/C, or O6-meG/T base pairs.
Similar articles
- Mismatch repair proteins as sensors of alkylation DNA damage.
Wang JY, Edelmann W. Wang JY, et al. Cancer Cell. 2006 Jun;9(6):417-8. doi: 10.1016/j.ccr.2006.05.013. Cancer Cell. 2006. PMID: 16766259 - Interactions of human mismatch repair proteins MutSalpha and MutLalpha with proteins of the ATR-Chk1 pathway.
Liu Y, Fang Y, Shao H, Lindsey-Boltz L, Sancar A, Modrich P. Liu Y, et al. J Biol Chem. 2010 Feb 19;285(8):5974-82. doi: 10.1074/jbc.M109.076109. Epub 2009 Dec 22. J Biol Chem. 2010. PMID: 20029092 Free PMC article. - Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase.
Stojic L, Mojas N, Cejka P, Di Pietro M, Ferrari S, Marra G, Jiricny J. Stojic L, et al. Genes Dev. 2004 Jun 1;18(11):1331-44. doi: 10.1101/gad.294404. Genes Dev. 2004. PMID: 15175264 Free PMC article. - Activation of ATR-related protein kinase upon DNA damage recognition.
Ma M, Rodriguez A, Sugimoto K. Ma M, et al. Curr Genet. 2020 Apr;66(2):327-333. doi: 10.1007/s00294-019-01039-w. Epub 2019 Oct 17. Curr Genet. 2020. PMID: 31624858 Free PMC article. Review. - ATR signalling: more than meeting at the fork.
Nam EA, Cortez D. Nam EA, et al. Biochem J. 2011 Jun 15;436(3):527-36. doi: 10.1042/BJ20102162. Biochem J. 2011. PMID: 21615334 Free PMC article. Review.
Cited by
- Accidental Encounter of Repair Intermediates in Alkylated DNA May Lead to Double-Strand Breaks in Resting Cells.
Fujii S, Fuchs RP. Fujii S, et al. Int J Mol Sci. 2024 Jul 26;25(15):8192. doi: 10.3390/ijms25158192. Int J Mol Sci. 2024. PMID: 39125763 Free PMC article. Review. - Molecular devices for high fidelity of DNA replication and gene expression.
Sekiguchi M. Sekiguchi M. Proc Jpn Acad Ser B Phys Biol Sci. 2006 Dec;82(8):278-96. doi: 10.2183/pjab.82.278. Epub 2006 Dec 2. Proc Jpn Acad Ser B Phys Biol Sci. 2006. PMID: 25792791 Free PMC article. Review. - Regulation of DNA Alkylation Damage Repair: Lessons and Therapeutic Opportunities.
Soll JM, Sobol RW, Mosammaparast N. Soll JM, et al. Trends Biochem Sci. 2017 Mar;42(3):206-218. doi: 10.1016/j.tibs.2016.10.001. Epub 2016 Nov 2. Trends Biochem Sci. 2017. PMID: 27816326 Free PMC article. Review. - Ascorbate acts as a highly potent inducer of chromate mutagenesis and clastogenesis: linkage to DNA breaks in G2 phase by mismatch repair.
Reynolds M, Stoddard L, Bespalov I, Zhitkovich A. Reynolds M, et al. Nucleic Acids Res. 2007;35(2):465-76. doi: 10.1093/nar/gkl1069. Epub 2006 Dec 14. Nucleic Acids Res. 2007. PMID: 17169990 Free PMC article. - The Promise of Poly(ADP-Ribose) Polymerase (PARP) Inhibitors in Gliomas.
Majd N, Yap TA, Yung WKA, de Groot J. Majd N, et al. J Immunother Precis Oncol. 2020 Nov 12;3(4):157-164. doi: 10.36401/JIPO-20-20. eCollection 2020 Nov. J Immunother Precis Oncol. 2020. PMID: 35665372 Free PMC article. Review.
References
- Abraham RT. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst.) 2004;3:883–887. - PubMed
- Berardini M, Mazurek A, Fishel R. The effect of O6-methylguanine DNA adducts on the adenosine nucleotide switch functions of hMSH2-hMSH6 and hMSH2-hMSH3. J. Biol. Chem. 2000;275:27851–27857. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous