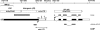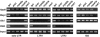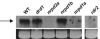Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis - PubMed (original) (raw)
Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis
Bruno Huettel et al. EMBO J. 2006.
Abstract
DRD1 is a SWI/SNF-like protein that cooperates with a plant-specific RNA polymerase, Pol IVb, to facilitate RNA-directed de novo methylation and silencing of homologous DNA. Screens to identify endogenous targets of this pathway in Arabidopsis revealed intergenic regions and plant genes located primarily in euchromatin. Many putative targets are near retrotransposon LTRs or other intergenic sequences that encode short RNAs, which might epigenetically regulate adjacent genes. Consistent with this, derepression of a solo LTR in drd1 and pol IVb mutants was accompanied by reduced cytosine methylation and transcriptional upregulation of neighboring sequences. The solo LTR and several other LTRs that flank reactivated targets are associated with euchromatic histone modifications but little or no H3K9 dimethylation, a hallmark of constitutive heterochromatin. By contrast, LTRs of retrotransposons that remain silent in the mutants despite reduced cytosine methylation lack euchromatic marks and have H3K9 dimethylation. We propose that DRD1 and Pol IVb establish a basal level of silencing that can potentially be reversed in euchromatin, and further reinforced in heterochromatin by other proteins that induce more stable modifications.
Figures
Figure 1
Bidirectional influence of a derepressed solo LTR in a drd1 mutant. Two transcripts were identified in both the SSH and cDNA-AFLP analyses: one matches a gene encoding the ribosomal protein RPL18C and the second contains part of the IG region together with the 3′ end of a truncated long interspersed element (LINE, a non-LTR retrotransposon) (gray boxes, SSH and cDNA-AFLP). The truncated LINE and the RPL18C gene are in the same orientation (horizontal white arrowheads) on chromosome 5 (coordinates at the top). The full-length transcripts (black bars), transcription start and termination sites, and transcript orientations (black arrows) were determined by 5′ and 3′ RACE. The sense RPL18C transcript initiates at the annotated start site whereas the antisense IG/LINE transcript initiates in the solo LTR, which served as the founding sequence element for a previously uncharacterized _Copia_-like retrotransposon family that we have named LTRCO (LTR/Copia) (Supplementary Table II). The antisense IG/LINE transcript terminates at a fortuitous polyadenylation site in the bottom DNA strand. Vertical white arrowheads in the solo LTR (which contains canonical U3, R, and U5 regions) indicate positions homologous to short RNAs identified in multiple parallel signature sequencing (Lu et al, 2005). The solo LTR was analyzed for histone modifications by ChIP (fine dotted line; primer positions as small arrows).
Figure 2
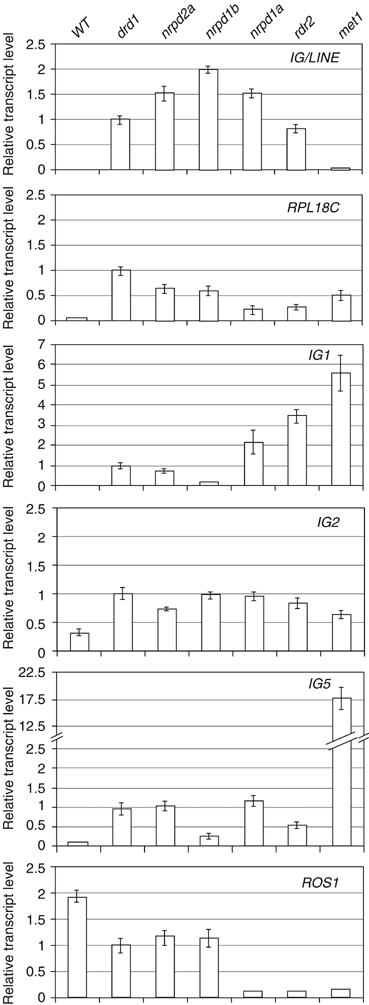
Differential expression of DRD1 targets. Real-time RT–PCR was used to validate transcripts that are upregulated (IG/LINE, RPL18C, IG1, IG2, IG5) or downregulated (ROS1) in the drd1 mutant as identified in SSH and/or cDNA-AFLP screens (Supplementary Table I). Relative transcript levels are shown in the indicated mutants: drd1, nrpd2a and nrpd1b (referred to collectively in the text as pol IVb mutants), nrpd1a, rdr2, and met1.
Figure 3
DNA methylation analysis. Cytosine methylation of the solo LTR, LTR1, LTR3 (all from the LTRCO family) and the Copia LTR acting as promoter for IG5 (Supplementary Figure 4C) in drd1, pol IVb (nrpd2a and nrpd1b), and met1 mutants was studied using enzymes sensitive to CG/CNG methylation (_Hpa_II), CNG methylation (_Msp_I), and CNN methylation (_Alu_I and _Dde_I). The solo LTR, LTR1, and LTR3 all have a single site for _Hpa_II/_Msp_I, but differ somewhat in the number of sites for _Dde_I and _Alu_I (Supplementary Figure 2A). Disappearance or reduced levels of a fragment after digestion with a given enzyme indicates loss of methylation at that site. The bottom panels show undigested input DNA.
Figure 4
Accumulation of LTR-derived short RNAs. LTR short RNAs from the LTRCO family of elements are detectable in wild-type plants and in drd1 and nrpd1b mutants, but not in nrpd2a, nrpd1a, and rdr2 mutants. The arrow indicates the position of a 24-nucleotide DNA oligonucleotide. Ethidium bromide staining of the major RNA species is shown as a loading control.
Figure 5
ChIP analysis of reactivated and silent DNA regions in drd1 and pol IVb mutants. (A) A heterochromatin control (At4g03770, a _Gypsy_-like retrotransposon) reacts with antibodies to H3K9me2 and H3K27me; (B, C) controls for euchromatin (At4g04040, a putative phosphofructokinase beta subunit and At5g23860, the tubulin protein TUB8) react with antibodies to H3K4me3 and acetylated H3, and weakly with H3K27me (Ac). The transgene α′ target promoter (D) and solo LTR (E) are derepressed in the drd1 and pol IVb (nrpd2a and nrpd1b) mutants. These sequences are enriched in the euchromatic marks (H3K4me3 and acetyl H3) and have H3K27me but little or no H3K9me2. For the transgene α′ promoter, the modifications are similar in wild-type (ST, DT) and mutant backgrounds. The solo LTR loses H3K27me and gains acetyl-H3 in the mutants. By contrast, LTR1 (F) and LTR3 (G), which remain silenced in drd1 and pol IVb mutants, are associated with H3K9me2 and enriched in H3K27me. The last lane in each panel shows a control using an antibody to unmodified histone H3. ST, transgenic line containing only the transgene α′ promoter-driven target gene; DT, transgenic line containing the silenced transgene α′ promoter-driven target gene and the unlinked silencer inverted repeat (Kanno et al, 2004, 2005b).
Figure 6
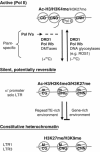
Model for reversible silencing of euchromatic genes by DRD1 and Pol IVb. Promoters of active genes transcribed by RNA polymerase II (Pol II) lack cytosine methylation and are enriched in euchromatic histone modifications (acetyl-H3 and H3K4me); some H3K27me may also be present. When short RNAs produced by Pol IVa are available, DRD1 and Pol IVb cooperate with DNA methyltransferases (DMTases) to induce de novo methylation (‘m') of cytosines in all sequence contexts in the region of RNA–DNA complementarity. Methylated, silenced promoters retain euchromatic marks and can show increased H3K27me. For euchromatic promoters that are sensitive to CNN methylation, such as the transgene α′ promoter and solo LTR, silencing can be reversed in dividing cells if the RNA trigger is removed, resulting in loss of CNN methylation. Cytosine methylation can also potentially be removed in nondividing cells through active demethylation by DNA glycosylase-domain-containing proteins such as ROS1, which might also involve the DRD1/Pol IVb machinery (Kanno et al, 2005a). Sequences in repeat or transposon (TE)-rich genomic environments, such as LTR1 and LTR3 of the LTRCO family, can be subjected to additional epigenetic modifications, including H3K9me and persistent C(N)G methylation (capital ‘M') to ensure stable silencing in constitutive heterochromatin. Sequences in gene-rich regions, such as the transgene α′ promoter and solo LTR, appear to be protected from the additional layer of epigenetic modifications, keeping them in a potentially reversible epigenetic state.
Similar articles
- RNA-directed DNA methylation and Pol IVb in Arabidopsis.
Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. Matzke M, et al. Cold Spring Harb Symp Quant Biol. 2006;71:449-59. doi: 10.1101/sqb.2006.71.028. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381327 Review. - RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants.
Huettel B, Kanno T, Daxinger L, Bucher E, van der Winden J, Matzke AJ, Matzke M. Huettel B, et al. Biochim Biophys Acta. 2007 May-Jun;1769(5-6):358-74. doi: 10.1016/j.bbaexp.2007.03.001. Epub 2007 Mar 12. Biochim Biophys Acta. 2007. PMID: 17449119 Review. - Arabidopsis RNA Polymerases IV and V Are Required To Establish H3K9 Methylation, but Not Cytosine Methylation, on Geminivirus Chromatin.
Jackel JN, Storer JM, Coursey T, Bisaro DM. Jackel JN, et al. J Virol. 2016 Jul 27;90(16):7529-7540. doi: 10.1128/JVI.00656-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27279611 Free PMC article. - The RNA silencing enzyme RNA polymerase v is required for plant immunity.
López A, Ramírez V, García-Andrade J, Flors V, Vera P. López A, et al. PLoS Genet. 2011 Dec;7(12):e1002434. doi: 10.1371/journal.pgen.1002434. Epub 2011 Dec 29. PLoS Genet. 2011. PMID: 22242006 Free PMC article. - DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV.
Zhang H, Ma ZY, Zeng L, Tanaka K, Zhang CJ, Ma J, Bai G, Wang P, Zhang SW, Liu ZW, Cai T, Tang K, Liu R, Shi X, He XJ, Zhu JK. Zhang H, et al. Proc Natl Acad Sci U S A. 2013 May 14;110(20):8290-5. doi: 10.1073/pnas.1300585110. Epub 2013 May 1. Proc Natl Acad Sci U S A. 2013. PMID: 23637343 Free PMC article.
Cited by
- Genome-Wide Analysis of DNA Demethylases in Land Plants and Their Expression Pattern in Rice.
Mao S, Xiao J, Zhao Y, Hou J, Li L. Mao S, et al. Plants (Basel). 2024 Jul 26;13(15):2068. doi: 10.3390/plants13152068. Plants (Basel). 2024. PMID: 39124186 Free PMC article. - Natural polymorphisms in ZMET2 encoding a DNA methyltransferase modulate the number of husk layers in maize.
Wang Z, Xia A, Wang Q, Cui Z, Lu M, Ye Y, Wang Y, He Y. Wang Z, et al. Plant Physiol. 2024 Jun 28;195(3):2129-2142. doi: 10.1093/plphys/kiae113. Plant Physiol. 2024. PMID: 38431291 Free PMC article. - Epigenetic regulation of plant immunity: from chromatin codes to plant disease resistance.
Xie SS, Duan CG. Xie SS, et al. aBIOTECH. 2023 Mar 17;4(2):124-139. doi: 10.1007/s42994-023-00101-z. eCollection 2023 Jun. aBIOTECH. 2023. PMID: 37581024 Free PMC article. Review. - The MOM1 complex recruits the RdDM machinery via MORC6 to establish de novo DNA methylation.
Li Z, Wang M, Zhong Z, Gallego-Bartolomé J, Feng S, Jami-Alahmadi Y, Wang X, Wohlschlegel J, Bischof S, Long JA, Jacobsen SE. Li Z, et al. Nat Commun. 2023 Jul 12;14(1):4135. doi: 10.1038/s41467-023-39751-4. Nat Commun. 2023. PMID: 37438334 Free PMC article. - Arms race between anti-silencing and RdDM in noncoding regions of transposable elements.
Sasaki T, Kato K, Hosaka A, Fu Y, Toyoda A, Fujiyama A, Tarutani Y, Kakutani T. Sasaki T, et al. EMBO Rep. 2023 Aug 3;24(8):e56678. doi: 10.15252/embr.202256678. Epub 2023 Jun 5. EMBO Rep. 2023. PMID: 37272687 Free PMC article.
References
- Breyne P, Dreesen R, Cannoot B, Rombaut D, Vandepoele K, Rombauts S, Vanderhaeghen R, Inze D, Zabeau M (2003) Quantitative cDNA-AFLP analysis for genome-wide expression studies. Mol Genet Genomics 269: 173–179 - PubMed
- Chan SW-L, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360 - PubMed
- Cigan AM, Unger-Wallace E, Haug-Collet K (2005) Transcriptional gene silencing as a tool for uncovering gene function in maize. Plant J 43: 929–940 - PubMed
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases