Using size exclusion chromatography-RPLC and RPLC-CIEF as two-dimensional separation strategies for protein profiling - PubMed (original) (raw)
Using size exclusion chromatography-RPLC and RPLC-CIEF as two-dimensional separation strategies for protein profiling
David C Simpson et al. Electrophoresis. 2006 Jul.
Abstract
Bottom-up proteomics (analyzing peptides that result from protein digestion) has demonstrated capability for broad proteome coverage and good throughput. However, due to incomplete sequence coverage, this approach is not ideally suited to the study of modified proteins. The modification complement of a protein can best be elucidated by analyzing the intact protein. 2-DE, typically coupled with the analysis of peptides that result from in-gel digestion, is the most frequently applied protein separation technique in MS-based proteomics. As an alternative, numerous column-based liquid phase techniques, which are generally more amenable to automation, are being investigated. In this work, the combination of size-exclusion chromatography (SEC) fractionation with RPLC-Fourier-transform ion cyclotron resonance (FTICR)-MS is compared with the combination of RPLC fractionation with CIEF-FTICR-MS for the analysis of the Shewanella oneidensis proteome. SEC-RPLC-FTICR-MS allowed the detection of 297 proteins, as opposed to 166 using RPLC-CIEF-FTICR-MS, indicating that approaches based on LC-MS provide better coverage. However, there were significant differences in the sets of proteins detected and both approaches provide a basis for accurately quantifying changes in protein and modified protein abundances.
Figures
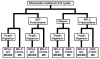
Figure 1
Schematic representation of the experimental pathway from S. oneidensis cell lysate to MS-analysis.
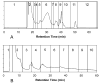
Figure 2
Chromatograms for (A) SEC fractionation and (B) RPLC fractionation of_S. oneidensis_ cell lysate; absorbance detection was performed at 280 nm; vertical lines indicate boundaries between fractions.
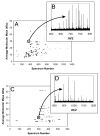
Figure 3
Comparison of (A) RPLC-FTICR-MS with (C) CIEF-FTICR-MS data, displayed as 2-D plots, for S. oneidensis cell lysate; CIEF sample preparation, in addition, required dialysis to reduce salt concentrations. 142 proteins were detected in the RPLC run, while 58 were detected in the CIEF run. (B) is the mass spectrum for the indicated spot in (A); the measured mass (average isotopic composition) for this spot was 29832.40 Da, which matches the mass of ribosomal protein L2 (SO0234) minus methionine at 9.6 ppm error. (D) is the mass spectrum for the indicated spot in (C); the measured mass (average isotopic composition) for this spot was 15127.80 Da, which matches the mass of hypothetical protein SO0691 at 0.3 ppm error.
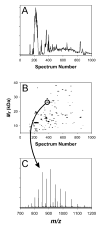
Figure 4
(A) TIC and (B) 2-D plot for SEC Fraction 5. (C) is the mass spectrum for the indicated spot in (B); the measured mass (average isotopic composition) for this spot was 26445.11 Da, which matches the mass of conserved hypothetical protein SO4719 minus a predicted signal peptide at 6.6 ppm error.
Figure 5
(A) TIC and (B) 2-D plot for RPLC Fraction 4. (C) is the mass spectrum for the indicated spot in (B); arrows highlight the peaks that constitute the indicated spot; the measured mass (average isotopic composition) for this spot was 10648.27 Da, which matches the mass of integration host factor beta subunit (SO2401) at 4.6 ppm error. (D) is an expansion of the 13+ peak that exemplifies the isotopic resolution achievable even for less intense peaks.
Similar articles
- Making broad proteome protein measurements in 1-5 min using high-speed RPLC separations and high-accuracy mass measurements.
Shen Y, Strittmatter EF, Zhang R, Metz TO, Moore RJ, Li F, Udseth HR, Smith RD, Unger KK, Kumar D, Lubda D. Shen Y, et al. Anal Chem. 2005 Dec 1;77(23):7763-73. doi: 10.1021/ac051257o. Anal Chem. 2005. PMID: 16316187 - Protein characterization by on-line capillary isoelectric focusing, reversed-phase liquid chromatography, and mass spectrometry.
Zhou F, Johnston MV. Zhou F, et al. Anal Chem. 2004 May 15;76(10):2734-40. doi: 10.1021/ac035446n. Anal Chem. 2004. PMID: 15144182 - FTICR mass spectrometry for qualitative and quantitative bioanalyses.
Page JS, Masselon CD, Smith RD. Page JS, et al. Curr Opin Biotechnol. 2004 Feb;15(1):3-11. doi: 10.1016/j.copbio.2004.01.002. Curr Opin Biotechnol. 2004. PMID: 15102459 Review. - Combining capillary electrophoresis with mass spectrometry for applications in proteomics.
Simpson DC, Smith RD. Simpson DC, et al. Electrophoresis. 2005 Apr;26(7-8):1291-305. doi: 10.1002/elps.200410132. Electrophoresis. 2005. PMID: 15765477 Review.
Cited by
- Fast, comprehensive two-dimensional liquid chromatography.
Stoll DR, Li X, Wang X, Carr PW, Porter SE, Rutan SC. Stoll DR, et al. J Chromatogr A. 2007 Oct 19;1168(1-2):3-43; discussion 2. doi: 10.1016/j.chroma.2007.08.054. Epub 2007 Aug 30. J Chromatogr A. 2007. PMID: 17888443 Free PMC article. Review. - Complete Characterization of Cardiac Myosin Heavy Chain (223 kDa) Enabled by Size-Exclusion Chromatography and Middle-Down Mass Spectrometry.
Jin Y, Wei L, Cai W, Lin Z, Wu Z, Peng Y, Kohmoto T, Moss RL, Ge Y. Jin Y, et al. Anal Chem. 2017 May 2;89(9):4922-4930. doi: 10.1021/acs.analchem.7b00113. Epub 2017 Apr 12. Anal Chem. 2017. PMID: 28366003 Free PMC article. - Ion Activation Methods for Peptides and Proteins.
Macias LA, Santos IC, Brodbelt JS. Macias LA, et al. Anal Chem. 2020 Jan 7;92(1):227-251. doi: 10.1021/acs.analchem.9b04859. Epub 2019 Nov 12. Anal Chem. 2020. PMID: 31665881 Free PMC article. Review. No abstract available. - Effective protein separation by coupling hydrophobic interaction and reverse phase chromatography for top-down proteomics.
Xiu L, Valeja SG, Alpert AJ, Jin S, Ge Y. Xiu L, et al. Anal Chem. 2014 Aug 5;86(15):7899-906. doi: 10.1021/ac501836k. Epub 2014 Jul 9. Anal Chem. 2014. PMID: 24968279 Free PMC article. - Comprehensive analysis of protein modifications by top-down mass spectrometry.
Zhang H, Ge Y. Zhang H, et al. Circ Cardiovasc Genet. 2011 Dec;4(6):711. doi: 10.1161/CIRCGENETICS.110.957829. Circ Cardiovasc Genet. 2011. PMID: 22187450 Free PMC article. Review.
References
- James P, Quadroni M, Carafoli E, Gonnet G. Biochem Biophys Res Comm. 1993;195:58–64. - PubMed
- Mann M, Hojrup P, Roepstorf P. Biol Mass Spectrom. 1993;22:338–345. - PubMed
- Pappin DJC, Hojrup P, Bleasby AJ. Curr Biol. 1993;3:327–332. - PubMed
- Yates JR, 3rd, Speicher S, Griffin PR, Hunkapiller T. Anal Biochem. 1993;214:397–408. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials