Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection - PubMed (original) (raw)
Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection
Tomoya Baba et al. Mol Syst Biol. 2006.
Abstract
We have systematically made a set of precisely defined, single-gene deletions of all nonessential genes in Escherichia coli K-12. Open-reading frame coding regions were replaced with a kanamycin cassette flanked by FLP recognition target sites by using a one-step method for inactivation of chromosomal genes and primers designed to create in-frame deletions upon excision of the resistance cassette. Of 4288 genes targeted, mutants were obtained for 3985. To alleviate problems encountered in high-throughput studies, two independent mutants were saved for every deleted gene. These mutants-the 'Keio collection'-provide a new resource not only for systematic analyses of unknown gene functions and gene regulatory networks but also for genome-wide testing of mutational effects in a common strain background, E. coli K-12 BW25113. We were unable to disrupt 303 genes, including 37 of unknown function, which are candidates for essential genes. Distribution is being handled via GenoBase (http://ecoli.aist-nara.ac.jp/).
Figures
Figure 1
Derivation of E. coli K-12 BW25113. Strain BD792, like MG1655, is a two-step descendent of ancestral E. coli K-12, EMG2, originally called WG1 (Bachmann, 1996; Hayashi et al, 2006; late BJ Bachmann, personal communication). Like its predecessor W1485F+ (Hayashi et al, 2006), BD792 has the rpoS396(Am) allele (codon 33, TAG (Am); unpublished data). Strain BW25113 was derived from BD792 in a series of steps involving generalized transduction and allele replacements, which included introducing the pseudoreversion rpoS (Q33) allele from MG1655 into a predecessor of BW25113 (Supplementary Table 1). The derivation of W3110 is shown in Figure 1 of accompanying manuscript (Hayashi et al, 2006).
Figure 2
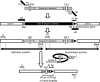
Primer design and construction of single-gene deletion mutants. Gene knockout primers have 20-nt 3′ ends for priming upstream (P1) and downstream (P2) of the FRT sites flanking the kanamycin resistance gene in pKD13 and 50-nt 5′ ends homologous to upstream (H1) and downstream (H2) chromosomal sequences for targeting the gene deletion (Supplementary Table 2). H1 includes the gene B (target) initiation codon. H2 includes codons for the six C-terminal residues, the stop codon, and 29-nt downstream. The same primer design with respect to gene B was used to target deletions regardless of whether gene B lies in an operon with genes A and C, as shown, or in different chromosomal arrangements. Novel junctions created between the resistance cassette and neighboring upstream (gene A) and downstream (gene C) sequences were verified by PCR with kanamycin (k1 or k2) and locus-specific (U or D) primers. Structures created after excision of the resistance gene are verified by PCR with neighboring gene-specific primers and by direct DNA sequencing of the region encompassing the H1-P1-FRT-P2-H2 scar to verify correct ones, as described elsewhere (Datsenko and Wanner, 2000). SD, Shine–Dalgarno ribosome binding sequence.
Figure 3
Structure of in-frame deletions. FLP-mediated excision of the FRT-flanked resistance gene is predicted to create a translatable scar sequence in-frame with the gene B target initiation codon and its C-terminal 18-nt coding region. Translation from the authentic gene B SD and start codon is expected to produce a 34-residue scar peptide with an N-terminal Met, 27 scar-specific residues, and six C-terminal, gene B-specific residues.
Figure 4
Mutagenesis of E. coli K-12 ORFs. See text.
Figure 5
COG classification of K-12 genes. See Supplementary Table 7.
Figure 6
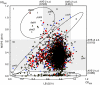
Profiling gene contribution for growth. Mutants of all 3985 genes in the Keio collection were grown 22 h in LB and 24 and 48 h in 0.4% glucose MOPS 2 mM Pi medium (Wanner, 1994). Maximum cell density values are plotted. Circled areas 1, 2, and 3 are discussed in text. Grayed areas show 2 × s.d. Groups labeled I–VII differ by more than 2 × s.d. Summary data are given in Table III. Additional time-course data are given in Supplementary Figure 1. Data are in given Supplementary Table 3. Major COG categories: blue diamond, information storage and processing; red square, cellular processes; yellow triangle, metabolism; black filled circle, poorly characterized; and black cross, no COG assignment.
Figure 7
Electron micrograph of E. coli K-12 by Melvin L Demphilis and Julius Adler. Republished with permission (Holden, 2002).
Figure 8
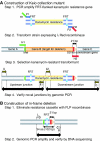
PCR gene replacement strategy. (A) Gene targeting fragment encoding kanamycin resistance with short homology extensions (H1 and H2) is generated by PCR by using priming sites P1 and P2 (Step 1). Gene targeting fragment is introduced into E. coli K-12 BW25113 expressing the Red recombinase from pKD46 (Step 2). Kanamycin-resistant transformants are selected (Step 3). Transformants are verified by PCR (Step 4). (B) Elimination of the resistance cassette by use of the FLP recombinase plasmid pCP20 is expected to leave behind a 102-nt ‘scar' encoding a 34-residue peptide (Step 1). The scar region is amplified and sequenced to be sure no mutations, especially 1-nt deletions (Datsenko and Wanner, 2000), were introduced (Step 2).
References
- Anderson RP, Roth JR (1977) Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol 31: 473–505 - PubMed
- Arifuzzaman M, Maeda M, Itoh A, Nishikata K, Takita C, Saito R, Ara T, Nakahigashi K, Hirai A, Tsuzuki K, Nakamura S, Altaf-Ul-Amin M, Oshima T, Baba T, Yamamoto N, Kawamura T, Ioka-Nakamichi T, Kitagawa M, Tomita M, Kanaya S, Wada C, Mori H (2006) Protein interaction networks of Escherichia coli K-12 (submitted) - PMC - PubMed
- Bachmann BJ (1996) Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In Escherichia coli and Salmonella typhimurium Cellular and Molecular Biology, Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low Jr KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds), 2 edn, pp 2460–2488. Washington, DC: ASM Press
- Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y (1997) The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases