Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water - PubMed (original) (raw)
Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water
Alice Layton et al. Appl Environ Microbiol. 2006 Jun.
Abstract
Bacteroides species are promising indicators for differentiating livestock and human fecal contamination in water because of their high concentration in feces and potential host specificity. In this study, a real-time PCR assay was designed to target Bacteroides species (AllBac) present in human, cattle, and equine feces. Direct PCR amplification (without DNA extraction) using the AllBac assay was tested on feces diluted in water. Fecal concentrations and threshold cycle were linearly correlated, indicating that the AllBac assay can be used to estimate the total amount of fecal contamination in water. Real-time PCR assays were also designed for bovine-associated (BoBac) and human-associated (HuBac) Bacteroides 16S rRNA genes. Assay specificities were tested using human, bovine, swine, canine, and equine fecal samples. The BoBac assay was specific for bovine fecal samples (100% true-positive identification; 0% false-positive identification). The HuBac assay had a 100% true-positive identification, but it also had a 32% false-positive rate with potential for cross-amplification with swine feces. The assays were tested using creek water samples from three different watersheds. Creek water did not inhibit PCR, and results from the AllBac assay were correlated with those from Escherichia coli concentrations (r2= 0.85). The percentage of feces attributable to bovine and human sources was determined for each sample by comparing the values obtained from the BoBac and HuBac assays with that from the AllBac assay. These results suggest that real-time PCR assays without DNA extraction can be used to quantify fecal concentrations and provide preliminary fecal source identification in watersheds.
Figures
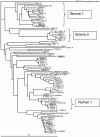
FIG. 1.
Phylogenetic dendrogram showing the relationship of cloned Bacteroides 16S rRNA gene sequences from different animal fecal sources. For each clone in this study, the first two letters represent the state (TN, Tennessee; PA, Pennsylvania; and TX, Texas), the next two letters represent the animal fecal source (Bo, cattle; Eq, horse; Av, chicken; Ca, dog; Sw, swine; and Hu, human), and the final number indicates the individual clone within the library. Sequences were aligned, and a bootstrap consensus tree was created using Clustal X (version 1.64b). The root was determined using the 16S rRNA gene sequence from Cytophaga fermentans (M58766) as an outgroup. References for cultured and uncultured Bacteroides or 16S rRNA gene sequences indicated on the tree were M58766 and M58762 (17); X83935, X83952, and X83953 (32); AB050110 (Y. Miyamoto, unpublished data) and AB021165 (27); and AF233400 and AF233408 (5). Plasmids containing the shaded sequences were used to determine the effect on sequence mismatches on real-time PCR assays in Fig. 2.
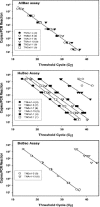
FIG. 2.
The effect of sequence mismatches on PCR amplification in real-time PCR assays. Serial dilutions of six different plasmids were performed to generate standard curves from 2.5 × 107 copies to 25 copies. Real-time PCR assays were performed as follows: top, AllBac assay with 0 sequence mismatches to all plasmids; middle, HuBac assay with 0 to 7 sequence mismatches; and bottom, BoBac assay with 0 to 14 sequence mismatches. Base pair mismatches between each plasmid and the primer and probe used in the real-time PCR assay are in parentheses in the legend boxes. Plasmids having more than seven mismatches to the primers and probe did not amplify.
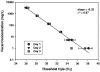
FIG. 3.
Threshold cycle measurements using the AllBac real time PCR assay in water samples containing bovine feces. Real-time PCR assays were performed on three separate days using triplicate samples for each dilution.
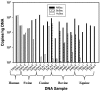
FIG. 4.
Concentration of ribosomal genes in DNA extracts from animal fecal samples determined by the AllBac, HuBac, and BoBac assays. For each sample, the first two letters represent the state (TN, Tennessee; PA, Pennsylvania; and TX, Texas) and the next two letters represent the animal fecal source (Bo, cattle; Eq, horse; Ca, dog; Sw, swine; and Hu, human).
FIG. 5.
Concentration of ribosomal genes in unextracted animal fecal samples determined by the AllBac, HuBac, and BoBac assays. For each sample, the first two letters represent the state (TN, Tennessee; PA, Pennsylvania; and TX, Texas) and the next two letters represent the animal fecal source (Bo, cattle; Eq, horse; Ca, dog; Sw, swine; and Hu, human).
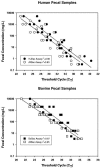
FIG. 6.
Comparison of AllBac and HuBac assays performed on serial dilutions from five individual human fecal samples (top), and the AllBac and BoBac assays run at 57°C on serial dilutions of six individual bovine fecal samples (bottom).
Similar articles
- Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater.
Okabe S, Okayama N, Savichtcheva O, Ito T. Okabe S, et al. Appl Microbiol Biotechnol. 2007 Mar;74(4):890-901. doi: 10.1007/s00253-006-0714-x. Epub 2006 Dec 1. Appl Microbiol Biotechnol. 2007. PMID: 17139508 - Quantitative identification of fecal water pollution sources by TaqMan real-time PCR assays using Bacteroidales 16S rRNA genetic markers.
Lee DY, Weir SC, Lee H, Trevors JT. Lee DY, et al. Appl Microbiol Biotechnol. 2010 Dec;88(6):1373-83. doi: 10.1007/s00253-010-2880-0. Epub 2010 Sep 25. Appl Microbiol Biotechnol. 2010. PMID: 20871990 - Lack of specificity for PCR assays targeting human Bacteroides 16S rRNA gene: cross-amplification with fish feces.
McLain JE, Ryu H, Kabiri-Badr L, Rock CM, Abbaszadegan M. McLain JE, et al. FEMS Microbiol Lett. 2009 Oct;299(1):38-43. doi: 10.1111/j.1574-6968.2009.01745.x. Epub 2009 Aug 1. FEMS Microbiol Lett. 2009. PMID: 19686344 - Discovering new indicators of fecal pollution.
McLellan SL, Eren AM. McLellan SL, et al. Trends Microbiol. 2014 Dec;22(12):697-706. doi: 10.1016/j.tim.2014.08.002. Epub 2014 Sep 5. Trends Microbiol. 2014. PMID: 25199597 Free PMC article. Review. - Ecophysiology and taxonomy of Bacteroides and related taxa.
Shah HN, Gharbia SE. Shah HN, et al. Clin Infect Dis. 1993 Jun;16 Suppl 4:S160-7. doi: 10.1093/clinids/16.supplement_4.s160. Clin Infect Dis. 1993. PMID: 7686781 Review.
Cited by
- Effect of maternal probiotic intervention on HPA axis, immunity and gut microbiota in a rat model of irritable bowel syndrome.
Barouei J, Moussavi M, Hodgson DM. Barouei J, et al. PLoS One. 2012;7(10):e46051. doi: 10.1371/journal.pone.0046051. Epub 2012 Oct 11. PLoS One. 2012. PMID: 23071537 Free PMC article. - Trends and determinants of gastric bacterial colonization of preterm neonates in a NICU setting.
Patel K, Konduru K, Patra AK, Chandel DS, Panigrahi P. Patel K, et al. PLoS One. 2015 Jul 1;10(7):e0114664. doi: 10.1371/journal.pone.0114664. eCollection 2015. PLoS One. 2015. PMID: 26132213 Free PMC article. - Efficacy and Gut Dysbiosis of Gentamicin-Intercalated Smectite as a New Therapeutic Agent against Helicobacter pylori in a Mouse Model.
Jeong SJ, Lee KH, Kim JH, Park SY, Song YG. Jeong SJ, et al. Antibiotics (Basel). 2020 Aug 10;9(8):502. doi: 10.3390/antibiotics9080502. Antibiotics (Basel). 2020. PMID: 32785101 Free PMC article. - Diet-driven microbial ecology underpins associations between cancer immunotherapy outcomes and the gut microbiome.
Simpson RC, Shanahan ER, Batten M, Reijers ILM, Read M, Silva IP, Versluis JM, Ribeiro R, Angelatos AS, Tan J, Adhikari C, Menzies AM, Saw RPM, Gonzalez M, Shannon KF, Spillane AJ, Velickovic R, Lazar AJ, Damania AV, Mishra AK, Chelvanambi M, Banerjee A, Ajami NJ, Wargo JA, Macia L, Holmes AJ, Wilmott JS, Blank CU, Scolyer RA, Long GV. Simpson RC, et al. Nat Med. 2022 Nov;28(11):2344-2352. doi: 10.1038/s41591-022-01965-2. Epub 2022 Sep 22. Nat Med. 2022. PMID: 36138151
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases