Biodiversity and classification of lactococcal phages - PubMed (original) (raw)
Biodiversity and classification of lactococcal phages
Hélène Deveau et al. Appl Environ Microbiol. 2006 Jun.
Abstract
For this study, an in-depth review of the classification of Lactococcus lactis phages was performed. Reference phages as well as unclassified phages from international collections were analyzed by stringent DNA-DNA hybridization studies, electron microscopy observations, and sequence analyses. A new classification scheme for lactococcal phages is proposed that reduces the current 12 groups to 8. However, two new phages (Q54 and 1706), which are unrelated to known lactococcal phages, may belong to new emerging groups. The multiplex PCR method currently used for the rapid identification of phages from the three main lactococcal groups (936, c2, and P335) was improved and tested against the other groups, none of which gave a PCR product, confirming the specificity of this detection tool. However, this method does not detect all members of the highly diverse P335 group. The lactococcal phages characterized here were deposited in the Félix d'Hérelle Reference Center for Bacterial Viruses and represent a highly diverse viral community from the dairy environment.
Figures
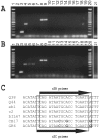
FIG. 1.
(A) Restriction profiles of lactococcal phages analyzed in this study. (B to G) Analysis of phage genomic DNAs by Southern hybridization, using the complete genomes of the following phages as probes: panel B, r1t; panel C, c2; panel D, P369; panel E, 949; panel F, Q54; panel G, 1706. Lanes 1 and 19, 1-kb DNA ladder (Invitrogen); lanes 2, phage bIL170 (genome digested with EcoRV); lanes 3, c2 (EcoRI); lanes 4, Q54 (EcoRI); lanes 5, GR6 (EcoRI); lanes 6, CB17 (EcoRI); lanes 7, ul36 (EcoRV); lanes 8, r1t (EcoRV); lanes 9, 1483 (EcoRV); lanes 10, T189 (EcoRV); lanes 11, P087 (EcoRV); lanes 12, 949 (EcoRV); lanes 13, bIL168 (EcoRV); lanes 14, 1706 (EcoRV); lanes 15, 1358 (EcoRI); lanes 16, KSY1 (AsnI); lanes 17, P369 (EcoRV); lanes 18, 1138 (EcoRV); lanes 20, BK5-T (EcoRV).
FIG. 2.
Identification of lactococcal phage species by multiplex PCR. (A) Three pairs of previously described primers (29) were used for PCR. (B) The same three sets of primers were used, except that the c2B primer was replaced by the c2C primer. Lanes 1 and 20, 100-bp DNA ladder (Invitrogen); lanes 2, negative control; lanes 3, bIL170; lanes 4, c2; lanes 5, GR6; lanes 6, CB17; lanes 7, Q54; lanes 8, r1t; lanes 9, 1483; lanes 10, ul36; lanes 11, T189; lanes 12, P087; lanes 13, 949; lanes 14, bIL168; lanes 15, 1706; lanes 16, 1358; lanes 17, KSY1; lanes 18, P369; lanes 19, 1138; lanes 21, BK5-T. (C) Analysis of the region covered by primers c2B and c2C in c2-like phages for which the gene coding for the major capsid protein is available in GenBank.
FIG. 3.
Alignment of the genetic maps of 10 completely sequenced P335-like phages (updated from reference 30). Deduced proteins sharing >60% amino acid identity are represented using the same colors and linked with gray shading when possible. Open reading frames with unique sequences are displayed in white. The dUTPase genes are identified by thick lines.
Similar articles
- Biodiversity of Lactococcus lactis bacteriophages in Polish dairy environment.
Szczepańska AK, Hejnowicz MS, Kołakowski P, Bardowski J. Szczepańska AK, et al. Acta Biochim Pol. 2007;54(1):151-8. Epub 2007 Mar 9. Acta Biochim Pol. 2007. PMID: 17311108 - Phage Biodiversity in Artisanal Cheese Wheys Reflects the Complexity of the Fermentation Process.
Mahony J, Moscarelli A, Kelleher P, Lugli GA, Ventura M, Settanni L, van Sinderen D. Mahony J, et al. Viruses. 2017 Mar 16;9(3):45. doi: 10.3390/v9030045. Viruses. 2017. PMID: 28300778 Free PMC article. - Multiplex PCR for detection and identification of lactococcal bacteriophages.
Labrie S, Moineau S. Labrie S, et al. Appl Environ Microbiol. 2000 Mar;66(3):987-94. doi: 10.1128/AEM.66.3.987-994.2000. Appl Environ Microbiol. 2000. PMID: 10698762 Free PMC article. - Species and type phages of lactococcal bacteriophages.
Jarvis AW, Fitzgerald GF, Mata M, Mercenier A, Neve H, Powell IB, Ronda C, Saxelin M, Teuber M. Jarvis AW, et al. Intervirology. 1991;32(1):2-9. doi: 10.1159/000150179. Intervirology. 1991. PMID: 1901837 Review. - Phages of dairy bacteria.
Brussow H. Brussow H. Annu Rev Microbiol. 2001;55:283-303. doi: 10.1146/annurev.micro.55.1.283. Annu Rev Microbiol. 2001. PMID: 11544357 Review.
Cited by
- Lytic infection of Lactococcus lactis by bacteriophages Tuc2009 and c2 triggers alternative transcriptional host responses.
Ainsworth S, Zomer A, Mahony J, van Sinderen D. Ainsworth S, et al. Appl Environ Microbiol. 2013 Aug;79(16):4786-98. doi: 10.1128/AEM.01197-13. Epub 2013 May 31. Appl Environ Microbiol. 2013. PMID: 23728817 Free PMC article. - Investigation of the relationship between lactococcal host cell wall polysaccharide genotype and 936 phage receptor binding protein phylogeny.
Mahony J, Kot W, Murphy J, Ainsworth S, Neve H, Hansen LH, Heller KJ, Sørensen SJ, Hammer K, Cambillau C, Vogensen FK, van Sinderen D. Mahony J, et al. Appl Environ Microbiol. 2013 Jul;79(14):4385-92. doi: 10.1128/AEM.00653-13. Epub 2013 May 10. Appl Environ Microbiol. 2013. PMID: 23666332 Free PMC article. - Characterization of Lactococcus lactis phage 949 and comparison with other lactococcal phages.
Samson JE, Moineau S. Samson JE, et al. Appl Environ Microbiol. 2010 Oct;76(20):6843-52. doi: 10.1128/AEM.00796-10. Epub 2010 Aug 27. Appl Environ Microbiol. 2010. PMID: 20802084 Free PMC article. - Isolation and Complete Genome Sequence of a Novel Marinobacter Phage B23.
Zhu M, Wang M, Jiang Y, You S, Zhao G, Liu Y, Yang Q, Liu Q, Liu Z, Gong Z, Shao H. Zhu M, et al. Curr Microbiol. 2018 Dec;75(12):1619-1625. doi: 10.1007/s00284-018-1568-z. Epub 2018 Sep 14. Curr Microbiol. 2018. PMID: 30218176 - Phages of non-dairy lactococci: isolation and characterization of ΦL47, a phage infecting the grass isolate Lactococcus lactis ssp. cremoris DPC6860.
Cavanagh D, Guinane CM, Neve H, Coffey A, Ross RP, Fitzgerald GF, McAuliffe O. Cavanagh D, et al. Front Microbiol. 2014 Jan 13;4:417. doi: 10.3389/fmicb.2013.00417. eCollection 2014 Jan 13. Front Microbiol. 2014. PMID: 24454309 Free PMC article.
References
- Ackermann, H.-W. 2001. Frequency of morphological phage descriptions in the year 2000. Brief review. Arch. Virol. 146:843-857. - PubMed
- Ackermann, H.-W. 2003. Bacteriophage observations and evolution. Res. Microbiol. 154:245-251. - PubMed
- Ackermann, H.-W., E. D. Cantor, A. W. Jarvis, J. Lembke, and J. A. Mayo. 1984. New species definitions in phages of gram-positive cocci. Intervirology 22:181-190. - PubMed
- Ackermann, H.-W., M. S. Dubow, A. W. Jarvis, L. A. Jones, V. N. Krylov, J. Maniloff, J. Rocourt, R. S. Safferman, J. Schneider, L. Seldin, T. Sozzi, P. R. Stewart, M. Werquin, and L. Wünsche. 1992. The species concept and its application to tailed phages. Arch. Virol. 124:69-82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous