New lineage of filamentous, spore-forming, gram-positive bacteria from soil - PubMed (original) (raw)
New lineage of filamentous, spore-forming, gram-positive bacteria from soil
Linda Cavaletti et al. Appl Environ Microbiol. 2006 Jun.
Abstract
A novel bacterial strain that was isolated from an Italian soil and was designated SOSP1-21T forms branched mycelia in solid and liquid media and has a filamentous morphology similar to that of some genera belonging to the Actinobacteria. Electron microscopy showed that this organism has a grape-like appearance, resulting from interlacing of spores originating from sporophoric hyphae. Ten strains that are morphologically related to SOSP1-21T were recovered from soil. Phylogenetic analyses of 16S rRNA gene segments confirmed the relatedness of these strains to SOSP1-21T and indicated that the newly isolated strains form separate clades in a deeply branching lineage. The closest matches for the 16S rRNA sequences of all the strains (around 79% identity) were matches with representatives of the Chloroflexi, although the affiliation with this division was not supported by high bootstrap values. The strains are mesophilic aerobic heterotrophs and are also capable of growing under microaerophilic conditions. They all stain gram positive. Strain SOSP1-21T contains ornithine, alanine, glutamic acid, serine, and glycine as the peptidoglycan amino acids. In addition, an unusual level of C16:1 2OH (30%) was found in the cellular fatty acids. The G+C content of SOSP1-21T genomic DNA is 53.9%, and MK-9(H2) was the only menaquinone detected. All these data suggest that SOSP1-21T and the related strains may constitute a new division of filamentous, spore-forming, gram-positive bacteria. We propose the name Ktedobacter racemifer gen. nov., sp. nov. for strain SOSP1-21T.
Figures
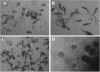
FIG. 1.
Light micrographs of strain SOSP1-21T. (a) Aerial morphology on an HSA5 plate after incubation for 4 weeks. Magnification, ×400. (b) Magnified (ca. ×2.5) image of the area in panel a indicated by the box, showing aerial hyphae and spherical spores. (c) Mycelial growth in a submerged culture after 3 days.
FIG. 2.
Appearance of strains on acidic ISP3 agar after 3 weeks. 1-21, strain SOSP1-21T; 1-85, strain SOSP1-85; 1-52, strain SOSP1-52; 1-9, strain SOSP1-9; 1-79, strain SOSP1-79; 1-1, strain SOSP1-1.
FIG. 3.
Light micrographs (obtained with a ULWD objective) of strains SOSP1-52 (A), SOSP1-1 (B), SOSP1-85 (C), and SOSP1-9 (D) on HSA5 plates after incubation for 4 weeks. Magnification, ×400.
FIG. 4.
FESEM of strain SOSP1-21T. Bars = 2 μm. See the text for details.
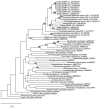
FIG. 5.
Maximum likelihood tree based on 1,202 aligned positions of the 16S rRNA gene. The tree was rooted using the 16S rRNA gene sequence from Methanococcus jannaschii (accession no. M59126) as the outgroup. The numbers at nodes are bootstrap values based on 100 replicated data sets; only values greater than 65 are shown. Scale bar = 10 inferred substitutions per 100 nucleotides.
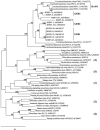
FIG. 6.
Maximum likelihood tree based on 1,168 aligned positions of the 16S rRNA gene. The tree was rooted using the Aquifex pyrophilus 16S rRNA sequence (accession no. M83548) as the outgroup. The numbers at nodes are bootstrap values based on 100 replicated data sets; only values greater than 65 are shown. Scale bar = 10 inferred substitutions per 100 nucleotides. The numbers in parentheses indicate the subphylum-level groups proposed by Hugenholtz and Stackebrandt (21), as follows: 1, “_Anaerolineae_”; 2, “_Dehalococcoidetes_”; 3, Chloroflexi; 5, Thermomicrobia. There are no cultivated representatives for lineage 4. The clades of highly related sequences are clades GER1 to GER3 for isolates and clade HAW1 for environmental clones (see the text for details).
Similar articles
- Reticulibacter mediterranei gen. nov., sp. nov., within the new family Reticulibacteraceae fam. nov., and Ktedonospora formicarum gen. nov., sp. nov., Ktedonobacter robiniae sp. nov., Dictyobacter formicarum sp. nov. and Dictyobacter arantiisoli sp. nov., belonging to the class Ktedonobacteria.
Yabe S, Zheng Y, Wang CM, Sakai Y, Abe K, Yokota A, Donadio S, Cavaletti L, Monciardini P. Yabe S, et al. Int J Syst Evol Microbiol. 2021 Jul;71(7). doi: 10.1099/ijsem.0.004883. Int J Syst Evol Microbiol. 2021. PMID: 34296987 - Dictyobacter aurantiacus gen. nov., sp. nov., a member of the family Ktedonobacteraceae, isolated from soil, and emended description of the genus Thermosporothrix.
Yabe S, Sakai Y, Abe K, Yokota A, Také A, Matsumoto A, Sugiharto A, Susilowati D, Hamada M, Nara K, Made Sudiana I, Otsuka S. Yabe S, et al. Int J Syst Evol Microbiol. 2017 Aug;67(8):2615-2621. doi: 10.1099/ijsem.0.001985. Epub 2017 Jul 31. Int J Syst Evol Microbiol. 2017. PMID: 28758628 - Planctomonas deserti gen. nov., sp. nov., a new member of the family Microbacteriaceae isolated from soil of the Taklamakan desert.
Liu SW, Li FN, Zheng HY, Qi X, Huang DL, Xie YY, Sun CH. Liu SW, et al. Int J Syst Evol Microbiol. 2019 Mar;69(3):616-624. doi: 10.1099/ijsem.0.003095. Epub 2018 Nov 29. Int J Syst Evol Microbiol. 2019. PMID: 30387709
Cited by
- Genome Features and Secondary Metabolites Biosynthetic Potential of the Class Ktedonobacteria.
Zheng Y, Saitou A, Wang CM, Toyoda A, Minakuchi Y, Sekiguchi Y, Ueda K, Takano H, Sakai Y, Abe K, Yokota A, Yabe S. Zheng Y, et al. Front Microbiol. 2019 Apr 26;10:893. doi: 10.3389/fmicb.2019.00893. eCollection 2019. Front Microbiol. 2019. PMID: 31080444 Free PMC article. - An integrated microbiological and electrochemical approach to determine distributions of Fe metabolism in acid mine drainage-induced "iron mound" sediments.
Leitholf IM, Fretz CE, Mahanke R, Santangelo Z, Senko JM. Leitholf IM, et al. PLoS One. 2019 Mar 26;14(3):e0213807. doi: 10.1371/journal.pone.0213807. eCollection 2019. PLoS One. 2019. PMID: 30913215 Free PMC article. - Contrasting bacterial communities in two indigenous Chionochloa (Poaceae) grassland soils in New Zealand.
Griffith JC, Lee WG, Orlovich DA, Summerfield TC. Griffith JC, et al. PLoS One. 2017 Jun 28;12(6):e0179652. doi: 10.1371/journal.pone.0179652. eCollection 2017. PLoS One. 2017. PMID: 28658306 Free PMC article. - The enigmatic SAR202 cluster up close: shedding light on a globally distributed dark ocean lineage involved in sulfur cycling.
Mehrshad M, Rodriguez-Valera F, Amoozegar MA, López-García P, Ghai R. Mehrshad M, et al. ISME J. 2018 Mar;12(3):655-668. doi: 10.1038/s41396-017-0009-5. Epub 2017 Dec 5. ISME J. 2018. PMID: 29208946 Free PMC article. - Analysis of microbial communities in heavy metals-contaminated soils using the metagenomic approach.
Hemmat-Jou MH, Safari-Sinegani AA, Mirzaie-Asl A, Tahmourespour A. Hemmat-Jou MH, et al. Ecotoxicology. 2018 Nov;27(9):1281-1291. doi: 10.1007/s10646-018-1981-x. Epub 2018 Sep 21. Ecotoxicology. 2018. PMID: 30242595
References
- Busti, E., L. Cavaletti, P. Monciardini, P. Schumann, M. Rohde, M. Sosio, and S. Donadio. Catenulispora acidiphila gen. nov., sp. nov., a novel mycelium-forming actinomycete and proposal of Catenulisporaceae fam. nov. Int. J. Syst. Evol. Microbiol., in press. - PubMed
- Busti, E., P. Monciardini, L. Cavaletti, R. Bamonte, A. Lazzarini, M. Sosio, and S. Donadio. 2006. Antibiotic producing ability by previously uncultured groups of actinomycetes. Microbiology 152:675-683. - PubMed
- Carnio, M. C., T. Stachelhaus, K. P. Francis, and S. Scherer. 2001. Pyridinyl polythiazole class peptide antibiotic micrococcin P1, secreted by foodborne Staphylococcus equorum WS2733, is biosynthesized nonribosomally. Eur. J. Biochem. 268:6390-6400. - PubMed
- Cavaletti, L., P. Monciardini, P. Schumann, M. Rohde, R. Bamonte, E. Busti, M. Sosio, and S. Donadio. Actinospica acidiphila gen. nov., sp. nov., and Actinospica robiniae gen. nov., sp. nov.; proposal for Actinospicaceae fam. nov. and Catenulisporinae subordo nov. in the order Actinomycetales. Int. J. Syst. Evol. Microbiol., in press. - PubMed
- Challis, G. L. 2005. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochemistry 6:601-611. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases