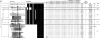Evaluation of the epidemiological relevance of variable-number tandem-repeat genotyping of Mycobacterium bovis and comparison of the method with IS6110 restriction fragment length polymorphism analysis and spoligotyping - PubMed (original) (raw)
Comparative Study
Evaluation of the epidemiological relevance of variable-number tandem-repeat genotyping of Mycobacterium bovis and comparison of the method with IS6110 restriction fragment length polymorphism analysis and spoligotyping
Caroline Allix et al. J Clin Microbiol. 2006 Jun.
Erratum in
- J Clin Microbiol. 2006 Sep;44(9):3471
Abstract
Sources of Mycobacterium bovis contamination remain unclear for many cases of animal and human disease. A major limitation is the lack of sufficiently informative or epidemiologically well evaluated molecular methods for typing. Here, we report an evaluation of a high-throughput method based on 29 mycobacterial interspersed repetitive unit-variable-number tandem-repeat (MIRU-VNTR) loci to genotype 127 M. bovis isolates from cattle from 77 different Belgian farms, representative of a nationwide collection obtained from 1995 to 2003. MIRU-VNTR stability was demonstrated by analyzing a series of 74 isolates in total, obtained from different animals from a single farm or from different farms with an identified epidemiological link. The genotyping results and the genotypic diversity (h) were compared with those obtained by IS6110 restriction fragment length polymorphism (RFLP) analysis and spoligotyping. Among 68 isolates with no known epidemiological link, MIRU-VNTR typing discriminated better than either RFLP analysis or spoligotyping, [corrected] taken individually (32 versus 16 and 17 genotypes; h = 0.91 versus 0.73 and 0.85, respectively) or in combination (32 versus 28 genotypes; h = 0.91 versus 0.92). Maximal resolution was already achieved with a subset of 9 loci. The observed congruence of the genetic relationships based on IS6110 RFLP analysis, spoligotyping, and MIRU-VNTR markers is consistent with a clonal population structure of M. bovis. These results support MIRU-VNTR typing as a convenient and discriminatory technique for analysis of the population structure of M. bovis in much greater detail and for addressing some still unresolved issues in the epidemiology of the pathogen.
Figures
FIG. 1.
IS_6110_ RFLP, spoligotyping, and MIRU-VNTR patterns of isolates obtained from different animals from a single farm. The dendrogram at the left is based on MIRU-VNTR profiles and was built by using the UPGMA algorithm, as described in Materials and Methods. Double alleles (4 and 7) detected in locus 3232 of two pooled samples (02011T1 and 02011T2) are boxed. a, a number (Nb) of >1 indicates a sample obtained by pooling tissues of n different animals from the same herd before mycobacteriological culture.
FIG. 2.
IS_6110_ RFLP, spoligotyping, and MIRU-VNTR patterns of isolates obtained from animals from different farms included in five distinct clusters with defined epidemiological links. The dendrogram at the left is based on MIRU-VNTR profiles and was built by using the UPGMA algorithm, as described in Materials and Methods. The single repeat change in locus ETR-A of sample 00059 compared to the same locus of the other isolates of cluster CE1 is boxed. a, a number (Nb) of >1 indicates a sample obtained by pooling tissues of n different animals from the same herd before mycobacteriological culture. See text and Fig. 3 for descriptions of farms and clusters.
FIG. 3.
Bovine tuberculosis transmission chain between different farms involving animals sold by one cattle trader. The cattle trader bought animals from farmer A and sold them to farmer B. In turn, animals from farm B were transferred to farm C. Samples from farms B and C and the cattle trader displayed completely identical genotypes (from the three genotyping methods) to that of isolate 00105T from farm A. Presumed clonal variants with a single band change in the IS_6110_ RFLP pattern (see boxed RFLP profiles for isolates 00059 and 00081T7) accompanied by a single repeat change in the ETR-A locus for isolate 00059 (see boxed allele) were found on farms A and D, respectively.
FIG. 4.
Congruence of genetic relationships between MIRU-VNTR typing, IS_6110_ RFLP, and spoligotyping. The dendrogram was constructed based on MIRU-VNTR genotypes using the neighbor-joining method, as described in Materials and Methods. The MIRU-VNTR tree was rooted using the Mycobacterium canetti clonal group C MIRU-VNTR genotype (11). The figure displays 37 different patterns among the isolates in panel 3, identified by the three genotyping methods. Boxed alleles correspond to MIRU-VNTR differences observed among samples within the G, H, J, and K lineages (see text). a, the number (Nb) of isolates with an identical genotype using the three genotyping methods. b, isolates with an identical IS_6110_ fingerprint within RFLP groups which were distinguished by MIRU-VNTR typing.
Similar articles
- Assessment of an optimized mycobacterial interspersed repetitive- unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis.
Oelemann MC, Diel R, Vatin V, Haas W, Rüsch-Gerdes S, Locht C, Niemann S, Supply P. Oelemann MC, et al. J Clin Microbiol. 2007 Mar;45(3):691-7. doi: 10.1128/JCM.01393-06. Epub 2006 Dec 27. J Clin Microbiol. 2007. PMID: 17192416 Free PMC article. - Sensitivities and specificities of spoligotyping and mycobacterial interspersed repetitive unit-variable-number tandem repeat typing methods for studying molecular epidemiology of tuberculosis.
Scott AN, Menzies D, Tannenbaum TN, Thibert L, Kozak R, Joseph L, Schwartzman K, Behr MA. Scott AN, et al. J Clin Microbiol. 2005 Jan;43(1):89-94. doi: 10.1128/JCM.43.1.89-94.2005. J Clin Microbiol. 2005. PMID: 15634955 Free PMC article. - [New era in molecular epidemiology of tuberculosis in Japan].
Takashima T, Iwamoto T. Takashima T, et al. Kekkaku. 2006 Nov;81(11):693-707. Kekkaku. 2006. PMID: 17154049 Review. Japanese. - Molecular epidemiology of bovine tuberculosis. I. Mycobacterium bovis genotyping.
Durr PA, Hewinson RG, Clifton-Hadley RS. Durr PA, et al. Rev Sci Tech. 2000 Dec;19(3):675-88. doi: 10.20506/rst.19.3.1241. Rev Sci Tech. 2000. PMID: 11107611 Review.
Cited by
- Professional Acquisition of M. bovis in Calabria Region (Southern Italy): A Challenging Case of Osteomyelitis in a Migrant Patient from Bulgaria.
Quirino A, Torti C, Strazzulla A, Nisticò S, Galati L, Barreca GS, Lamberti AG, Berardelli G, Pacciarini M, Gasparini G, Pisani V, Gambardella A, Liberto MC, Focà A. Quirino A, et al. Case Rep Infect Dis. 2015;2015:794715. doi: 10.1155/2015/794715. Epub 2015 Jul 14. Case Rep Infect Dis. 2015. PMID: 26257970 Free PMC article. - Molecular Characterization and Drug Susceptibility of Mycobacterium Bovis Isolated from Cattle in Xinjiang, China.
Ou XC, Xu F, Zhou Y, Tian LL, Zeng QY, Fan WX, Zhao YL. Ou XC, et al. Chin Med J (Engl). 2018 Dec 20;131(24):3017-3019. doi: 10.4103/0366-6999.247204. Chin Med J (Engl). 2018. PMID: 30539923 Free PMC article. No abstract available. - Evaluation of the new advanced 15-loci MIRU-VNTR genotyping tool in Mycobacterium tuberculosis molecular epidemiology studies.
Alonso-Rodríguez N, Martínez-Lirola M, Herránz M, Sanchez-Benitez M, Barroso P; INDAL-TB group; Bouza E, García de Viedma D. Alonso-Rodríguez N, et al. BMC Microbiol. 2008 Feb 24;8:34. doi: 10.1186/1471-2180-8-34. BMC Microbiol. 2008. PMID: 18339198 Free PMC article. - First insights into the genetic diversity of Mycobacterium tuberculosis isolates from HIV-infected Mexican patients and mutations causing multidrug resistance.
Lopez-Alvarez R, Badillo-Lopez C, Cerna-Cortes JF, Castillo-Ramirez I, Rivera-Gutierrez S, Helguera-Repetto AC, Aguilar D, Hernandez-Pando R, Samper S, Gonzalez-y-Merchand JA. Lopez-Alvarez R, et al. BMC Microbiol. 2010 Mar 17;10:82. doi: 10.1186/1471-2180-10-82. BMC Microbiol. 2010. PMID: 20236539 Free PMC article. - Usefulness of mycobacterial interspersed repetitive-unit locus PCR amplification in rapid diagnosis of Beijing lineage strain infection among pediatric tuberculosis patients.
Liu R, Xing L, Peng Z, Zhang Y, Zhu C, Yang Z. Liu R, et al. J Clin Microbiol. 2011 Feb;49(2):712-4. doi: 10.1128/JCM.01694-10. Epub 2010 Dec 1. J Clin Microbiol. 2011. PMID: 21123538 Free PMC article.
References
- Alito, A., N. Morcillo, S. Scipioni, A. Dolmann, M. I. Romano, A. Cataldi, and D. van Soolingen. 1999. The IS6110 restriction fragment length polymorphism in particular multidrug-resistant Mycobacterium tuberculosis strains may evolve too fast for reliable use in outbreak investigation. J. Clin. Microbiol. 37:788-791. - PMC - PubMed
- Allix, C., P. Supply, and M. Fauville-Dufaux. 2004. Utility of fast mycobacterial interspersed repetitive unit-variable number tandem repeat genotyping in clinical mycobacteriological analysis. Clin. Infect. Dis. 39:783-789. - PubMed
- Ayele, W. Y., S. D. Neill, J. Zinsstag, M. G. Weiss, and I. Pavlik. 2004. Bovine tuberculosis: an old disease but a new threat to Africa. Int. J. Tuberc. Lung Dis. 8:924-937. - PubMed
- Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources