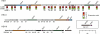A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus - PubMed (original) (raw)
. 2006 Jun 9;312(5779):1526-30.
doi: 10.1126/science.1128393.
Marianne E Cuff, Stefan Raunser, Aimee Shen, Min Zhou, Casey A Gifford, Andrew L Goodman, Grazyna Joachimiak, Claudia L Ordoñez, Stephen Lory, Thomas Walz, Andrzej Joachimiak, John J Mekalanos
Affiliations
- PMID: 16763151
- PMCID: PMC2800167
- DOI: 10.1126/science.1128393
A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus
Joseph D Mougous et al. Science. 2006.
Abstract
Bacterial pathogens frequently use protein secretion to mediate interactions with their hosts. Here we found that a virulence locus (HSI-I) of Pseudomonas aeruginosa encodes a protein secretion apparatus. The apparatus assembled in discrete subcellular locations and exported Hcp1, a hexameric protein that forms rings with a 40 angstrom internal diameter. Regulatory patterns of HSI-I suggested that the apparatus functions during chronic infections. We detected Hcp1 in pulmonary secretions of cystic fibrosis (CF) patients and Hcp1-specific antibodies in their sera. Thus, HSI-I likely contributes to the pathogenesis of P. aeruginosa in CF patients. HSI-I-related loci are widely distributed among bacterial pathogens and may play a general role in mediating host interactions.
Figures
Fig. 1
Overview of P. aeruginosa HSI genes and reciprocal regulation of HSI-I by RetS and LadS. Conserved hypothetical HSI ORFs not discussed in the text (white) and ORFs that lie within the predicted HSI operons (black) are labeled with their genome annotation ORF number [assigned on the basis of (7)]. Predicted paralogous ORFs with prior characterization and those characterized in this study are colored consistently in each locus. The boxed insert shows the position of the hcp2/vgrG2 locus encoded elsewhere in the genome. The boxes beneath HSI-I genes summarize transcriptional profiling data from two prior studies (4, 5). The asterisks denote measurements that meet the statistical significance threshold defined in each study.
Fig. 2
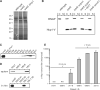
P. aeruginosa HSI-I is required for secretion of Hcp1 and is active in cystic fibrosis infections. (A) Hcp1 is hypersecreted by Δ_retS_. SDS–polyacrylamide gel electrophoresis analysis of concentrated culture supernatants from various P. aeruginosa strains. The arrow highlights the position of secreted Hcp1 in Δ_retS_. (B) Immunoblot analysis of HSI-I–dependent secretion of Hcp1-V. In addition to the genetic alterations indicated, each strain contains hcp1-V. Equal quantities of cell (C) and supernatant (S) fractions were probed with antibodies specific for the β-subunit of RNA polymerase (RNAP) and the VSV-G epitope. (C) Immunoblot analysis of Hcp1 secretion by control strains and a panel of CF patient clinical isolates. (D) Immunoblot analysis of Hcp1 in sputum from CF patients (upper blot). Sputum sample 002-6 is from a CF patient not infected with P. aeruginosa; sputum sample 180-8 is from a CF patient infected with two P. aeruginosa strains that do not secrete Hcp1 (lower blot); and sputum sample 195-1 is from a CF patient infected with a P. aeruginosa strain that actively secretes Hcp1 and a second that does not (lower blot). (E) ELISA analysis of sera from CF patients for antibody response against Hcp1.
Fig. 3
ClpV1 is not involved in thermotolerance and localizes to discrete foci in a manner dependent on IcmF and Hcp1. (A) Thermotolerance assay of P. aeruginosa strains bearing a clpB or clpV1 deletion. Cell viability before and after a 25-min heat pulse at 55°C was determined by colony-forming units. The thermotolerance of each strain was normalized relative to wild-type. (B and C) Immunoblot analysis of Hcp1-V secretion by ClpV1 and the AAA-1 mutant ClpV1E310A (B) and in Δ_retS clpV1-gfp_ (C). (D) Fluorescence microscopy of indicated strains also bearing clpV1-gfp. TMA-DPH is a membrane dye used to highlight the outline of the cells.
Fig. 4
Hcp1 forms a hexameric ring with a large internal diameter. (A) Ribbon representation of the Hcp1 monomer colored by secondary structure: β strands, red; α helices, blue; and loops, green. (B) Top view of a ribbon representation of the crystallographic Hcp1 hexamer. The individual subunits are colored differently to highlight their organization. (C) Edge-on view of the Hcp1 hexamer shown in (B). (D) Electron microscopy and single-particle analysis of Hcp1. Electron micrograph of Hcp1 negatively stained with 0.75% (w/v) uranyl formate. Scale bar, 100 nm. (Inset) (Left) Representative class averages and (right) the same averages after six-fold symmetrization. Inset scale bar, 10 nm. (E) Sequence conservation analysis of Hcp1. An alignment of 107 Hcp proteins in 43 Gram-negative bacteria was used to plot the relative degree of conservation at each amino acid on the surface of Hcp1 (see methods in supporting online material). Conservation is indicated by color, where red residues are highly conserved and white residues are poorly conserved.
Similar articles
- Cystic Fibrosis Lung Function Decline after Within-Host Evolution Increases Virulence of Infecting Pseudomonas aeruginosa.
Jorth P, Durfey S, Rezayat A, Garudathri J, Ratjen A, Staudinger BJ, Radey MC, Genatossio A, McNamara S, Cook DA, Aitken ML, Gibson RL, Yahr TL, Singh PK. Jorth P, et al. Am J Respir Crit Care Med. 2021 Mar 1;203(5):637-640. doi: 10.1164/rccm.202007-2735LE. Am J Respir Crit Care Med. 2021. PMID: 33137262 Free PMC article. No abstract available. - Genetic features of Pseudomonas aeruginosa isolates from cystic fibrosis patients compared with those of isolates from other origins.
Lanotte P, Watt S, Mereghetti L, Dartiguelongue N, Rastegar-Lari A, Goudeau A, Quentin R. Lanotte P, et al. J Med Microbiol. 2004 Jan;53(Pt 1):73-81. doi: 10.1099/jmm.0.05324-0. J Med Microbiol. 2004. PMID: 14663109 - The Small RNA ErsA Plays a Role in the Regulatory Network of Pseudomonas aeruginosa Pathogenicity in Airway Infections.
Ferrara S, Rossi A, Ranucci S, De Fino I, Bragonzi A, Cigana C, Bertoni G. Ferrara S, et al. mSphere. 2020 Oct 14;5(5):e00909-20. doi: 10.1128/mSphere.00909-20. mSphere. 2020. PMID: 33055260 Free PMC article. - Regulation of alginate gene expression in Pseudomonas aeruginosa.
Zielinski NA, Roychoudhury S, Chakrabarty AM. Zielinski NA, et al. Methods Enzymol. 1994;235:493-502. doi: 10.1016/0076-6879(94)35165-1. Methods Enzymol. 1994. PMID: 8057921 Review. No abstract available. - Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors.
Deretic V, Schurr MJ, Boucher JC, Martin DW. Deretic V, et al. J Bacteriol. 1994 May;176(10):2773-80. doi: 10.1128/jb.176.10.2773-2780.1994. J Bacteriol. 1994. PMID: 8188579 Free PMC article. Review. No abstract available.
Cited by
- Unique substrates secreted by the type VI secretion system of Francisella tularensis during intramacrophage infection.
Bröms JE, Meyer L, Sun K, Lavander M, Sjöstedt A. Bröms JE, et al. PLoS One. 2012;7(11):e50473. doi: 10.1371/journal.pone.0050473. Epub 2012 Nov 20. PLoS One. 2012. PMID: 23185631 Free PMC article. - Roles of Hcp family proteins in the pathogenesis of the porcine extraintestinal pathogenic Escherichia coli type VI secretion system.
Peng Y, Wang X, Shou J, Zong B, Zhang Y, Tan J, Chen J, Hu L, Zhu Y, Chen H, Tan C. Peng Y, et al. Sci Rep. 2016 May 27;6:26816. doi: 10.1038/srep26816. Sci Rep. 2016. PMID: 27229766 Free PMC article. - Secretome analysis of Vibrio cholerae type VI secretion system reveals a new effector-immunity pair.
Altindis E, Dong T, Catalano C, Mekalanos J. Altindis E, et al. mBio. 2015 Mar 10;6(2):e00075. doi: 10.1128/mBio.00075-15. mBio. 2015. PMID: 25759499 Free PMC article. - Structural basis for type VI secretion effector recognition by a cognate immunity protein.
Li M, Le Trong I, Carl MA, Larson ET, Chou S, De Leon JA, Dove SL, Stenkamp RE, Mougous JD. Li M, et al. PLoS Pathog. 2012;8(4):e1002613. doi: 10.1371/journal.ppat.1002613. Epub 2012 Apr 12. PLoS Pathog. 2012. PMID: 22511866 Free PMC article. - A metaproteomics approach to elucidate host and pathogen protein expression during catheter-associated urinary tract infections (CAUTIs).
Lassek C, Burghartz M, Chaves-Moreno D, Otto A, Hentschker C, Fuchs S, Bernhardt J, Jauregui R, Neubauer R, Becher D, Pieper DH, Jahn M, Jahn D, Riedel K. Lassek C, et al. Mol Cell Proteomics. 2015 Apr;14(4):989-1008. doi: 10.1074/mcp.M114.043463. Epub 2015 Feb 11. Mol Cell Proteomics. 2015. PMID: 25673765 Free PMC article.
References
- Yahr TL, Greenberg EP. Mol. Cell. 2004;16:497. - PubMed
- Goodman AL, et al. Dev. Cell. 2004;7:745. - PubMed
- Ventre I, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM074942/GM/NIGMS NIH HHS/United States
- R01 AI026289/AI/NIAID NIH HHS/United States
- AI26289/AI/NIAID NIH HHS/United States
- U54 GM074942-04S2/GM/NIGMS NIH HHS/United States
- R01 AI021451/AI/NIAID NIH HHS/United States
- P50 GM062414/GM/NIGMS NIH HHS/United States
- P50 GM062414-02/GM/NIGMS NIH HHS/United States
- GM62414/GM/NIGMS NIH HHS/United States
- R37 AI021451/AI/NIAID NIH HHS/United States
- AI21451/AI/NIAID NIH HHS/United States
- U54 GM074942/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases