Reconstructing genetic ancestry blocks in admixed individuals - PubMed (original) (raw)
Reconstructing genetic ancestry blocks in admixed individuals
Hua Tang et al. Am J Hum Genet. 2006 Jul.
Abstract
A chromosome in an individual of recently admixed ancestry resembles a mosaic of chromosomal segments, or ancestry blocks, each derived from a particular ancestral population. We consider the problem of inferring ancestry along the chromosomes in an admixed individual and thereby delineating the ancestry blocks. Using a simple population model, we infer gene-flow history in each individual. Compared with existing methods, which are based on a hidden Markov model, the Markov-hidden Markov model (MHMM) we propose has the advantage of accounting for the background linkage disequilibrium (LD) that exists in ancestral populations. When there are more than two ancestral groups, we allow each ancestral population to admix at a different time in history. We use simulations to illustrate the accuracy of the inferred ancestry as well as the importance of modeling the background LD; not accounting for background LD between markers may mislead us to false inferences about mixed ancestry in an indigenous population. The MHMM makes it possible to identify genomic blocks of a particular ancestry by use of any high-density single-nucleotide-polymorphism panel. One application of our method is to perform admixture mapping without genotyping special ancestry-informative-marker panels.
Figures
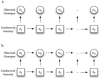
Figure 1.
Graphical representation of an HMM (a) and an MHMM (b).
Figure 2.
Estimation of two-marker haplotype frequency estimation. Unphased genotype data in 50 individuals were simulated on the basis of chromosome 22 haplotypes of the CEU individuals genotyped in the HapMap project. Each plot can be viewed as a two-dimensional histogram, in which the _X_-axis represents the true haplotype frequency, and the _Y_-axis represents the corresponding estimated frequencies. The intensity at each pixel indicates the height of the histogram, or the number of marker pairs whose true haplotype frequency is at the _X_-coordinate while the estimated haplotype frequency is at the _Y_-coordinate. a, Naive haplotype frequency estimates. Both allele frequencies and haplotype frequencies are estimated from a small sample of individuals. b, Augmented haplotype frequency estimates. Haplotype frequencies were estimated from same set of individuals as in panel a, but allele frequencies were estimated from a larger sample.
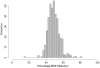
Figure 3.
Estimated admixing time, τ, of 400 simulated individuals. Red circles represent the MLE under the MHMM; blue triangles represent the MLE under the HMM by use of the same genotype data. True times are 25, 10, and 25, indicated with a yellow square. Some jitter is added to the MLEs to aid visualization.
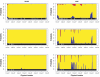
Figure 4.
Ancestry for a simulated admixed individual. The _Y_-axis represents the posterior probability that one allele is derived from a specific ancestry; the _X_-axis indicates the physical locations of the markers. Top, True ancestral states. Middle, MHMM estimates. Bottom, HMM estimates.
Figure 5.
Comparison of percentage reduction in MSE. Percentage reduction for individual n is defined as
(MSE HMM _n_-MSE MHMM n)/MSE HMM n
.
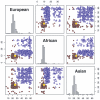
Figure 6.
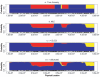
Estimated ancestry for a Han Chinese individual from Beijing. The _Y_-axis represents the posterior probability that one allele is derived from a specific ancestry; the _X_-axis indicates the physical locations of markers. Markers were sampled at an average spacing of 30 kb (top panels), 6 kb (middle panels), and 3 kb (bottom panels), which approximated the density of a 100K SNP chip, approximated the density of a 500K SNP chip, and used all HapMap SNPs, respectively. Left panels, MHMM correctly infers Asian ancestry (yellow) at most markers. Right panels, HMM assigns considerable probability of European ancestry (blue) or African ancestry (red) in several regions.
Figure 7.
Estimated ancestry for a simulated individual with asymmetric admixing history. The _Y_-axis represents the posterior probability that one allele is derived from a specific ancestry; the _X_-axis indicates the physical locations of markers. a, True ancestry along the paternal and the maternal chromosomes. The paternal chromosome was generated assuming
τ=(25,25,25)
and
π=(0.4,0.4,0.2)
, whereas the maternal chromosome was generated assuming
τ=(2,2,2)
and
π=(0.75,0.125,0.125)
. b, Posterior ancestry estimates at the MLE of τ. c, Posterior ancestry estimates under the assumption
τ=(2,2,2)
. d, Posterior ancestry estimates under the assumption
τ=(50,50,50)
.
Similar articles
- Inferring ancestries efficiently in admixed populations with linkage disequilibrium.
Bercovici S, Geiger D. Bercovici S, et al. J Comput Biol. 2009 Aug;16(8):1141-50. doi: 10.1089/cmb.2009.0105. J Comput Biol. 2009. PMID: 19645595 - An ancestry informative marker panel design for individual ancestry estimation of Hispanic population using whole exome sequencing data.
Wang LJ, Zhang CW, Su SC, Chen HH, Chiu YC, Lai Z, Bouamar H, Ramirez AG, Cigarroa FG, Sun LZ, Chen Y. Wang LJ, et al. BMC Genomics. 2019 Dec 30;20(Suppl 12):1007. doi: 10.1186/s12864-019-6333-6. BMC Genomics. 2019. PMID: 31888480 Free PMC article. - Mapping asthma-associated variants in admixed populations.
Mersha TB. Mersha TB. Front Genet. 2015 Sep 29;6:292. doi: 10.3389/fgene.2015.00292. eCollection 2015. Front Genet. 2015. PMID: 26483834 Free PMC article. Review. - A Continuous Correlated Beta Process Model for Genetic Ancestry in Admixed Populations.
Gompert Z. Gompert Z. PLoS One. 2016 Mar 11;11(3):e0151047. doi: 10.1371/journal.pone.0151047. eCollection 2016. PLoS One. 2016. PMID: 26966908 Free PMC article. - Analyses of genetic ancestry enable key insights for molecular ecology.
Gompert Z, Buerkle CA. Gompert Z, et al. Mol Ecol. 2013 Nov;22(21):5278-94. doi: 10.1111/mec.12488. Epub 2013 Sep 19. Mol Ecol. 2013. PMID: 24103088 Review.
Cited by
- Inference of Locus-Specific Population Mixtures from Linked Genome-Wide Allele Frequencies.
Reyna-Blanco CS, Caduff M, Galimberti M, Leuenberger C, Wegmann D. Reyna-Blanco CS, et al. Mol Biol Evol. 2024 Jul 3;41(7):msae137. doi: 10.1093/molbev/msae137. Mol Biol Evol. 2024. PMID: 38958167 Free PMC article. - Power comparison of admixture mapping and direct association analysis in genome-wide association studies.
Qin H, Zhu X. Qin H, et al. Genet Epidemiol. 2012 Apr;36(3):235-43. doi: 10.1002/gepi.21616. Epub 2012 Mar 28. Genet Epidemiol. 2012. PMID: 22460597 Free PMC article. - Dating the age of admixture via wavelet transform analysis of genome-wide data.
Pugach I, Matveyev R, Wollstein A, Kayser M, Stoneking M. Pugach I, et al. Genome Biol. 2011;12(2):R19. doi: 10.1186/gb-2011-12-2-r19. Epub 2011 Feb 25. Genome Biol. 2011. PMID: 21352535 Free PMC article. - Effect of genetic divergence in identifying ancestral origin using HAPAA.
Sundquist A, Fratkin E, Do CB, Batzoglou S. Sundquist A, et al. Genome Res. 2008 Apr;18(4):676-82. doi: 10.1101/gr.072850.107. Epub 2008 Mar 18. Genome Res. 2008. PMID: 18353807 Free PMC article. - Imputation of exome sequence variants into population- based samples and blood-cell-trait-associated loci in African Americans: NHLBI GO Exome Sequencing Project.
Auer PL, Johnsen JM, Johnson AD, Logsdon BA, Lange LA, Nalls MA, Zhang G, Franceschini N, Fox K, Lange EM, Rich SS, O'Donnell CJ, Jackson RD, Wallace RB, Chen Z, Graubert TA, Wilson JG, Tang H, Lettre G, Reiner AP, Ganesh SK, Li Y. Auer PL, et al. Am J Hum Genet. 2012 Nov 2;91(5):794-808. doi: 10.1016/j.ajhg.2012.08.031. Epub 2012 Oct 25. Am J Hum Genet. 2012. PMID: 23103231 Free PMC article.
References
Web Resource
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials