Genomic instability due to V(D)J recombination-associated transposition - PubMed (original) (raw)
Genomic instability due to V(D)J recombination-associated transposition
Yeturu V R Reddy et al. Genes Dev. 2006.
Abstract
The first step in assembling immunoglobulin and T-cell receptors by V(D)J recombination has similarities to transposon excision. The excised transposon-like element then integrates into DNA targets at random in vitro, but whether this activity significantly threatens the genomic integrity of its host has been unclear. Here, we recover examples where the putative transposon associated with V(D)J recombination integrated into the genome of a pre-B-cell line. Transposition accounted for a surprisingly high proportion (one-third) of integrations, while most of the remaining events had parallels to other aberrant V(D)J recombination pathways linked to oncogenic translocation. In total, transposition occurred approximately once every 50,000 V(D)J recombinations. Transposition may thus contribute significantly to genomic instability.
Figures
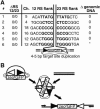
Figure 1.
Recovery of integrations of a transposon-like fragment. (A) The assay for recovering potential transposon integrations is described. (Boxes) Coding sequence; (filled triangle) 12-type recombination signal (12-RS); (open triangle) 23-type recombination signal (23-RS); (puror) intact gene for puromycin resistance; (zeorGFP) fusion gene that confers zeocin resistance and green fluorescence. In B and C, DNA from subclones resistant to puromycin and zeocin after induction of V(D)J recombination (numbered as in Table 2) were digested with EcoRI (cuts once 5′ of the puror gene) and PstI (cuts only in flanking chromosomal DNA), Southern blotted, and analyzed with a probe from the puror gene (B) or zeorGFP gene (C). DNA from the parental line (P) and the initial clone with unrearranged substrate (clone A) are included for comparison. The mobility of molecular weight markers (in kilobase pairs) are shown at the right of each panel.
Figure 2.
Integrations consistent with transpositions. (A) For each of the identified integration events (#, numbered as in Table 2) we report the number of base pairs deleted from 12-RS and 23-RS ends of the zeorGFP fragment (ΔRS 12/23), the chromosome where the fragment integrated (Ch.), a brief description of genomic DNA immediately flanking 12-RS ends and 23-RS ends, and the amount of genomic DNA between integration flanks (Δ genomic DNA). The sequences of flanking target site duplications are in bold. At the bottom is a graphic summarizing the typical integration structure. (*) As described in detail in footnote c of Table 2, the repetitive nature of sequences flanking integration #6 did not allow for unambiguous location of flanks within the region. (B) Pathway for integration by transposition. Ovals represent RAG proteins.
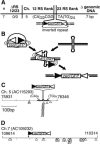
Figure 3.
Integration into an inverted repeat. (A) The structure of integration #7 is summarized as in Figure 2A. (B) Pathway for integration by transposition into a hairpin-forming sequence. Ovals represent RAG proteins. (C) A 415-bp chromosome 5 fragment that contained the targets for cellular integration #7 is represented as a line. The termini of the cloned fragment are defined by nucleotide numbers that map to the sequence in accession AC115293. The approximate location of (CA)N and (TG)N repeats within this fragment are noted by thick lines (see also footnote d of Table 2). The locations of cellular integration sites are noted above the line, while the locations of in vitro-defined integrations are noted with open triangles below the line. (D) Seven-hundred base pairs of a chromosome 7 fragment (from AC109232) that contained the target for cellular integration #8 are represented as in C. The site of integration of the 12-RS of the zeorGFP fragment is noted above the line. The 23-RS flank could not be located in this region (see footnote e of Table 2).
Figure 4.
Other nontargeted integrations. The structures of integration #8–11 are summarized as in Figures 2A and 3A. A possible inserted nucleotide (nontemplated) is noted in lowercase. (*) Sequences flanking the 23-RS for #8 and #9 were derived from Moloney Murine leukemia virus genes, and could not be located near the 12-RS flanks. (See also footnote e of Table 2.)
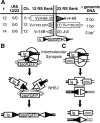
Figure 5.
Integration by intermolecular V(D)J recombination. (A) The structure of integrations #12–14 are summarized as in Figures 2, 3, 4. Rectangles represent flanking immunoglobulin (Ig) locus coding segments, and triangles represent flanking Ig locus recombination signals. The numbers in parentheses within these symbols refer to the base pairs of the genomic sequence deleted relative to the site of RAG protein cleavage. (*) Integration #14 is the product of two recombinations, as described in Supplementary Figure 2. B and C describe how intermolecular recombination can similarly lead to both integration of the zeorGFP fragment (B) as well as translocations between receptor loci and oncogene (onc) loci (C). Ovals represent RAG proteins, while boxes represent NHEJ proteins.
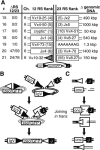
Figure 6.
Integration by end donation. (A) The structure of integrations #15–21 are summarized as in Figures 2, 3, 4, 5. Rectangles represent flanking Ig locus coding segments, and triangles represent flanking Ig locus recombination signals. The numbers in parentheses within these symbols refer to the base pairs of the genomic sequence deleted relative to the site of RAG protein cleavage. (*) Sequence flanks a cryptic 23-type signal within the Vκ region: CACtGTG (23 bp) AgAAAAACC (nucleotides that match consensus RS in capital letters). B and C describe how end donation can similarly lead to both integration of the zeorGFP fragment (B) as well as translocations between receptor loci and onc loci (C). Boxes represent NHEJ proteins.
Similar articles
- V(D)J recombination: how to tame a transposase.
Brandt VL, Roth DB. Brandt VL, et al. Immunol Rev. 2004 Aug;200:249-60. doi: 10.1111/j.0105-2896.2004.00161.x. Immunol Rev. 2004. PMID: 15242410 Review. - Transposition of hAT elements links transposable elements and V(D)J recombination.
Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL. Zhou L, et al. Nature. 2004 Dec 23;432(7020):995-1001. doi: 10.1038/nature03157. Nature. 2004. PMID: 15616554 - New concepts in the regulation of an ancient reaction: transposition by RAG1/RAG2.
Chatterji M, Tsai CL, Schatz DG. Chatterji M, et al. Immunol Rev. 2004 Aug;200:261-71. doi: 10.1111/j.0105-2896.2004.00167.x. Immunol Rev. 2004. PMID: 15242411 Review. - In vivo reinsertion of excised episomes by the V(D)J recombinase: a potential threat to genomic stability.
Vanura K, Montpellier B, Le T, Spicuglia S, Navarro JM, Cabaud O, Roulland S, Vachez E, Prinz I, Ferrier P, Marculescu R, Jäger U, Nadel B. Vanura K, et al. PLoS Biol. 2007 Mar;5(3):e43. doi: 10.1371/journal.pbio.0050043. PLoS Biol. 2007. PMID: 17298184 Free PMC article. - The taming of a transposon: V(D)J recombination and the immune system.
Jones JM, Gellert M. Jones JM, et al. Immunol Rev. 2004 Aug;200:233-48. doi: 10.1111/j.0105-2896.2004.00168.x. Immunol Rev. 2004. PMID: 15242409 Review.
Cited by
- HIV‑1 integrase inhibitors targeting various DDE transposases: Retroviral integration versus RAG‑mediated recombination (Review).
Mușat MG, Nițulescu GM, Surleac M, Tsatsakis A, Spandidos DA, Margină D. Mușat MG, et al. Mol Med Rep. 2019 Dec;20(6):4749-4762. doi: 10.3892/mmr.2019.10777. Epub 2019 Oct 30. Mol Med Rep. 2019. PMID: 31702817 Free PMC article. Review. - Chromosomal reinsertion of broken RSS ends during T cell development.
Curry JD, Schulz D, Guidos CJ, Danska JS, Nutter L, Nussenzweig A, Schlissel MS. Curry JD, et al. J Exp Med. 2007 Oct 1;204(10):2293-303. doi: 10.1084/jem.20070583. Epub 2007 Sep 4. J Exp Med. 2007. PMID: 17785508 Free PMC article. - RAG: a recombinase diversified.
Matthews AG, Oettinger MA. Matthews AG, et al. Nat Immunol. 2009 Aug;10(8):817-21. doi: 10.1038/ni.1776. Epub 2009 Jul 21. Nat Immunol. 2009. PMID: 19621044 Free PMC article. Review. - Initial stages of V(D)J recombination: the organization of RAG1/2 and RSS DNA in the postcleavage complex.
Grundy GJ, Ramón-Maiques S, Dimitriadis EK, Kotova S, Biertümpfel C, Heymann JB, Steven AC, Gellert M, Yang W. Grundy GJ, et al. Mol Cell. 2009 Jul 31;35(2):217-27. doi: 10.1016/j.molcel.2009.06.022. Mol Cell. 2009. PMID: 19647518 Free PMC article. - Aberrantly resolved RAG-mediated DNA breaks in Atm-deficient lymphocytes target chromosomal breakpoints in cis.
Mahowald GK, Baron JM, Mahowald MA, Kulkarni S, Bredemeyer AL, Bassing CH, Sleckman BP. Mahowald GK, et al. Proc Natl Acad Sci U S A. 2009 Oct 27;106(43):18339-44. doi: 10.1073/pnas.0902545106. Epub 2009 Oct 9. Proc Natl Acad Sci U S A. 2009. PMID: 19820166 Free PMC article.
References
- Agrawal A., Eastman Q.M., Schatz D.G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. - PubMed
- Bogue M.A., Wang C., Zhu C., Roth D.B. V(D)J recombination in Ku86-deficient mice: Distinct effects on coding, signal, and hybrid joint formation. Immunity. 1997;7:37–47. - PubMed
- Chen Y.Y., Wang L.C., Huang M.S., Rosenberg N. An active v-abl protein tyrosine kinase blocks immunoglobulin light-chain gene rearrangement. Genes & Dev. 1994;8:688–697. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials