The Saccharomyces cerevisiae rev6-1 mutation, which inhibits both the lesion bypass and the recombination mode of DNA damage tolerance, is an allele of POL30, encoding proliferating cell nuclear antigen - PubMed (original) (raw)
The Saccharomyces cerevisiae rev6-1 mutation, which inhibits both the lesion bypass and the recombination mode of DNA damage tolerance, is an allele of POL30, encoding proliferating cell nuclear antigen
Hengshan Zhang et al. Genetics. 2006 Aug.
Abstract
The rev6-1 allele was isolated in a screen for mutants deficient for UV-induced reversion of the frameshift mutation his4-38. Preliminary testing showed that the rev6-1 mutant was substantially deficient for UV-induced reversion of arg4-17 and ilv1-92 and markedly UV sensitive. Unlike other REV genes, which encode DNA polymerases and an associated subunit, REV6 has been found to be identical to POL30, which encodes proliferating cell nuclear antigen (PCNA), the subunit of the homotrimeric sliding clamp, in which the rev6-1 mutation produces a G178S substitution. This substitution appears to abolish all DNA damage-tolerance activities normally carried out by the RAD6/RAD18 pathway, including translesion replication by DNA polymerase zeta/Rev1 and DNA polymerase eta, and the error-free, recombination-dependent component of this pathway, but has little effect on the growth rate, suggesting that G178S may prevent ubiquitination of lysine 164 in PCNA. We also find that rev6-1 mutation can be fully complemented by a centromere-containing, low copy-number plasmid carrying POL30, despite the presumed occurrence in the mutant of sliding clamp assemblies that contain between one and three G178S PCNA monomers as well as the fully wild-type species.
Figures
Figure 1.—
Frequencies of UV-induced reversion of arg4-17 (A, C, and E) and percentage of survival following UV irradiation (B, D, and F) in REV6, rev6-1, _rev1_Δ, _rev3_Δ, _rad30_Δ, and _pol32_Δ strains and in _rev6-1 rev1_Δ, _rev6-1 rev3_Δ, _rev6-1 rad30_Δ, and _rev6-1 pol32_Δ double mutants. A–F: ▪, REV6+; *, rev6-1. Additionally, in A and B: ○, _rad30_Δ; ▿, _pol32_Δ; •, _rev1_Δ; ▾, _rev3_Δ. In C and D: ○, _rev6-1 rev1_Δ; ▿, _rev6-1 pol32_Δ ; •, _rev6-1 rad30_Δ; ▾, _rev6-1 rev3_Δ. In E and F: ○, rev6-1 + YCplac33 POL30; ▾, rev6-1 + YCplac33. Data are the average of two or three replicate experiments.
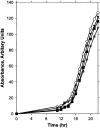
Figure 2.—
Growth rate monitored by turbidity (arbitrary absorbance units) and viable cell titer in REV6+ and rev6-1 strains transformed with either YCplac33 POL30 or YCplac33. ▪, REV6; *, rev6-1; ○, rev6-1 with YCplac33 POL30; ▾, rev6-1 with YCplac33. Data are the average of three experiments.
Similar articles
- Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae.
Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Haracska L, et al. Mol Cell Biol. 2004 May;24(10):4267-74. doi: 10.1128/MCB.24.10.4267-4274.2004. Mol Cell Biol. 2004. PMID: 15121847 Free PMC article. - The translesion DNA polymerases Pol ζ and Rev1 are activated independently of PCNA ubiquitination upon UV radiation in mutants of DNA polymerase δ.
Tellier-Lebegue C, Dizet E, Ma E, Veaute X, Coïc E, Charbonnier JB, Maloisel L. Tellier-Lebegue C, et al. PLoS Genet. 2017 Dec 27;13(12):e1007119. doi: 10.1371/journal.pgen.1007119. eCollection 2017 Dec. PLoS Genet. 2017. PMID: 29281621 Free PMC article. - Error-free DNA-damage tolerance in Saccharomyces cerevisiae.
Xu X, Blackwell S, Lin A, Li F, Qin Z, Xiao W. Xu X, et al. Mutat Res Rev Mutat Res. 2015 Apr-Jun;764:43-50. doi: 10.1016/j.mrrev.2015.02.001. Epub 2015 Feb 16. Mutat Res Rev Mutat Res. 2015. PMID: 26041265 Review. - Control of PCNA deubiquitylation in yeast.
Gallego-Sánchez A, Conde F, San Segundo P, Bueno A. Gallego-Sánchez A, et al. Biochem Soc Trans. 2010 Feb;38(Pt 1):104-9. doi: 10.1042/BST0380104. Biochem Soc Trans. 2010. PMID: 20074044 Review.
Cited by
- A novel function of DNA polymerase zeta regulated by PCNA.
Northam MR, Garg P, Baitin DM, Burgers PM, Shcherbakova PV. Northam MR, et al. EMBO J. 2006 Sep 20;25(18):4316-25. doi: 10.1038/sj.emboj.7601320. Epub 2006 Sep 7. EMBO J. 2006. PMID: 16957771 Free PMC article. - Solution X-ray scattering combined with computational modeling reveals multiple conformations of covalently bound ubiquitin on PCNA.
Tsutakawa SE, Van Wynsberghe AW, Freudenthal BD, Weinacht CP, Gakhar L, Washington MT, Zhuang Z, Tainer JA, Ivanov I. Tsutakawa SE, et al. Proc Natl Acad Sci U S A. 2011 Oct 25;108(43):17672-7. doi: 10.1073/pnas.1110480108. Epub 2011 Oct 17. Proc Natl Acad Sci U S A. 2011. PMID: 22006297 Free PMC article. - PrimPol-dependent single-stranded gap formation mediates homologous recombination at bulky DNA adducts.
Piberger AL, Bowry A, Kelly RDW, Walker AK, González-Acosta D, Bailey LJ, Doherty AJ, Méndez J, Morris JR, Bryant HE, Petermann E. Piberger AL, et al. Nat Commun. 2020 Nov 17;11(1):5863. doi: 10.1038/s41467-020-19570-7. Nat Commun. 2020. PMID: 33203852 Free PMC article. - The Many Roles of PCNA in Eukaryotic DNA Replication.
Boehm EM, Gildenberg MS, Washington MT. Boehm EM, et al. Enzymes. 2016;39:231-54. doi: 10.1016/bs.enz.2016.03.003. Epub 2016 Apr 19. Enzymes. 2016. PMID: 27241932 Free PMC article. Review.
References
- Gibbs, P. E. M., and C. W. Lawrence, 1995. Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. J. Mol. Biol. 251: 229–236. - PubMed
- Gibbs, P. E. M., J. MacDonald, R. Woodgate and C. W. Lawrence, 2005. The relative roles in vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 protein and Pol 32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct, and T-T cis-syn cyclobutane dimer. Genetics 169: 575–582. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous