Characterization of the primary starch utilization operon in the obligate anaerobe Bacteroides fragilis: Regulation by carbon source and oxygen - PubMed (original) (raw)
Characterization of the primary starch utilization operon in the obligate anaerobe Bacteroides fragilis: Regulation by carbon source and oxygen
Cheryl Spence et al. J Bacteriol. 2006 Jul.
Abstract
The opportunistic pathogen Bacteroides fragilis is a commensal organism in the large intestine, where it utilizes both dietary and host-derived polysaccharides as a source of carbon and energy. In this study, a four-gene operon required for starch utilization was identified. The operon also was found to be oxygen responsive and thus was designated osu for oxygen-induced starch utilization. The first three genes in the operon were predicted to encode outer membrane proteins involved in starch binding, and a fourth gene, osuD, encoded an amylase involved in starch hydrolysis. Insertional mutation of the osuA gene (Omega osuA) resulted in the inability to utilize starch or glycogen and an insertional mutation into the osuD gene (Omega osuD) was severely impaired for growth on starch media. Transcriptional studies indicated that maltose, maltooligosaccharides, and starch were inducers of osu expression and that maltose was the strongest inducer. A transcriptional activator of osuABCD, OsuR, was identified and found to mediate maltose induction. The Omega osuA and Omega osuD mutants were able to grow on maltose but not starch, whereas a mutation in osuR abolished growth on both substrates, indicating that additional genes under the control of OsuR are needed for maltose utilization. The osuABCD operon also was induced by exposure to oxygen and was shown to be part of the oxidative stress response important for aerotolerance of B. fragilis. Transcriptional analyses showed that osuA was induced 20-fold by oxygen, but OsuR was not required for this activation. Analysis of osu mutants suggested that expression of the operon was important for survival during oxygen exposure but not to hydrogen peroxide stress.
Figures
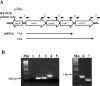
FIG. 1.
Functional map and operon structure of the osu locus. A. Functional map derived from base pairs 3709740 to 3720756 of the B. fragilis 638R genome sequence. The approximate sizes of osu mRNA transcripts are shown by arrows below the map and RT-PCR primer sets 1 through 5 are shown above the map. B. RT-PCR analysis of the osuABCD operon. Agarose gel showing RT-PCR products resulting from primer pairs shown in panel A. Lane designations are as follows: Mw, 1-kb molecular mass standard; lane 1, primer set 1; lane 2, primer set 2; lane 3, primer set 3; lane 4, primer set 4; lane 5, primer set 5. Lanes 6 and 7 were positive controls using primer sets 1 and 5, respectively, in PCRs with wild-type strain 638R DNA.
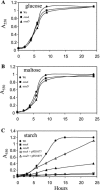
FIG. 2.
Growth of osu mutant strains (osuA and osuD) in SDM supplemented with glucose, maltose, or starch. Wt, wild type. A. SDM supplemented with 0.3% glucose. B. SDM supplemented with 0.3% maltose. C. SDM supplemented with 0.5% starch. Data are presented as the means of three independent experiments performed in triplicate.
FIG. 3.
Amylase activity of the B. fragilis osu mutant strains on starch azure plates. Ten microliters of an overnight BHIS culture of wild-type strain 638R (W+), IB367 (Ω_osuA_), and IB371 (Ω_osuD_) were spotted onto starch azure plates and incubated at 37°C. Starch azure plates contained 0.5% xylose, 0.3% starch, and 0.4% starch azure to allow for growth of the mutant strains.
FIG. 4.
Autoradiographs of Northern blots probed with internal fragments of osuA or osuD. Total RNA was isolated from mid-log-phase cultures of strain 638R grown anaerobically in SDM supplemented with either 0.3% glucose (G) or 0.5% starch (S) as the sole carbon/energy source. All lanes contain 30 μg of total RNA. Approximate sizes of the hybridizing transcripts were extrapolated from a molecular mass ladder in an adjacent lanes.
FIG. 5.
Role of secondary structure in the expression of osuD. A. Sequence of the intergenic region between osuC and osuD. The inverted repeat is shown by the arrows over the sequence and the 19-bp deletion disrupting the structure is shown by the dashed line under the sequence. B. Phosphorimager image of Northern blot analysis of wild-type 638R (W+) and the mutant IB442 (with the 19-bp deletion) (Δ19 bp) grown on SDM with starch as the sole carbon/energy source. The probe was an internal osuA gene fragment. The arrows to the right of the autoradiograph show the location of the 7- and 9-kb mRNA species. C. Amylase activity on starch azure plates. Ten microliters of overnight cultures grown in BHIS were spotted on starch azure plates as described in the legend to Fig. 3. Incubation was for 24 h at 37°C.
FIG. 6.
Induction of osuA and osuD expression by maltooligosaccharides in the wild-type strain 638R. Relative expression was determined by real-time RT-PCR during anaerobic growth in SDM supplemented with glucose (G1), maltose (G2), maltotriose (G3), maltopentaose (G5), maltoheptaose (G7) or starch (STA). Real-time RT-PCR results were normalized to the amount of 16S rRNA in each sample and then expressed as _n_-fold induction relative to the expression during growth in glucose medium. Data are presented as the means from three independent experiments.
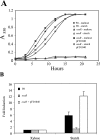
FIG. 7.
Role of OsuR in growth and gene expression. Wt, wild type (638R); osuR, osuR mutant. A. Effect of osuR mutation on growth in SDM supplemented with starch or maltose. B. Effect of starch on the expression of osuA in wild-type and osuR mutant strains. Relative expression levels were determined during anaerobic growth in a xylose-containing basal medium supplemented with or without 0.5% starch. Results are expressed as _n_-fold induction relative to expression levels in basal xylose medium without starch. Strains were grown anaerobically to mid-log phase and then 0.5% starch was added to induce osuABCD expression. Total RNA was isolated 1 h after starch addition and expression of osuA was analyzed by real-time RT-PCR. Data are presented as the means from three independent experiments.
FIG. 8.
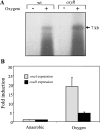
Oxygen-induced expression of the osu operon. A. Autoradiograph of Northern blot probed with internal osuA gene fragment. Total RNA was isolated from mid-log-phase cultures of strains 638R (wild type [wt]) and IB298 (oxyR) grown anaerobically in BHIS (−) or stressed by aerobic incubation for 1 h (+). All lanes contain 30 μg of total RNA. Approximate size of the hybridizing transcript, shown in kb, was extrapolated from a molecular mass ladder in an adjacent lane. B. Effect of oxygen stress on induction of osuA and osuD expression in the wild-type strain. Relative expression levels were determined by real-time RT-PCR for both anaerobic growth and oxygen exposure. Wild-type strain 638R was grown anaerobically to mid-log phase in BHIS and then exposed to oxygen for 1 h. Total RNA was isolated, and the expression of osuA and osuD was analyzed, with the results expressed as _n_-fold induction upon exposure to oxygen relative to anaerobic expression. Data are presented as the means from three independent experiments.
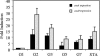
FIG. 9.
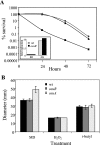
Effect of osu on survival during oxidative stress. A. Survival of wild-type (wt)(•), Ω_osuA_ (▪) and osuR (▴) strains following exposure to atmospheric oxygen. Mid-log-phase cells (A_550 of 0.35) grown in BHIS were exposed to oxygen by shaking at 250 rpm in air at 37°C. Viable cell counts were determined over 72 h. Inset, real-time RT-PCR analysis of osuA expression during oxygen exposure (ox) or under anaerobic conditions (an). The relative n_-fold increase in expression is compared to anaerobic conditions, which were normalized to 1. B. Sensitivity of Ω_osuA and osuR mutant strains to oxidizing agents. Disk diffusion assays were used to compare wild-type (638R), osuR (IB393), and Ω_osuA (IB367) strains. Results are reported as the diameter of growth inhibition around filters treated with the following oxidizing agents: 3% menadione (MD), 3% hydrogen peroxide (H2O2), 0.5% _t_-butyl hydroperoxide (_t_-butyl). Values are the means from three independent experiments performed in triplicate.
FIG. 10.
Identification of the osuABCD operon transcription initiation site. By use of 5′ RACE analysis, the transcription start site was located 42 bp upstream from the translation start site of OsuA during anaerobic growth conditions with starch as the sole carbon source. During oxygen stress transcription was initiated 44 bp from the translational start site. Upstream from the start sites, a conserved inverted repeat putative binding site for the LacI type regulator family is shown in bold with arrows over the sequence. The consensus cre binding sequence for CcpA is shown below the conserved LacI binding site; W, A or T; N, any nucleotide; ST, starch. Lines below and above the sequence indicate the putative −7 and −33 promoter consensus sequences.
References
- Baughn, A. D., and M. H. Malamy. 2004. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427:441-444. - PubMed
- Bayley, D. P., E. R. Rocha, and C. J. Smith. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 193:149-154. - PubMed
- Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. - PubMed
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases