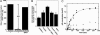Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases - PubMed (original) (raw)
Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases
William C Hallows et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Silent Information Regulator 2 (Sir2) enzymes (or sirtuins) are NAD(+)-dependent deacetylases that modulate gene silencing, aging and energy metabolism. Previous work has implicated several transcription factors as sirtuin targets. Here, we investigated whether mammalian sirtuins could directly control the activity of metabolic enzymes. We demonstrate that mammalian Acetyl-CoA synthetases (AceCSs) are regulated by reversible acetylation and that sirtuins activate AceCSs by deacetylation. Site-specific acetylation of mouse AceCS1 on Lys-661 was identified by using mass spectrometry and a specific anti-acetyl-AceCS antibody. SIRT1 was the only member of seven human Sir2 homologues capable of deacetylating AceCS1 in cellular coexpression experiments. SIRT1 expression also led to a pronounced increase in AceCS1-dependent fatty-acid synthesis from acetate. Using purified enzymes, only SIRT1 and SIRT3 exhibited high catalytic efficiency against acetylated AceCS1. In mammals, two AceCSs have been identified: cytoplasmic AceCS1 and mitochondrial AceCS2. Because SIRT3 is localized to the mitochondria, we investigated whether AceCS2 also might be regulated by acetylation, and specifically deacetylated by mitochondrial SIRT3. AceCS2 was completely inactivated upon acetylation and was rapidly reactivated by SIRT3 deacetylation. Lys-635 of mouse AceCS2 was identified as the targeted residue. Using reversible acetylation to modulate enzyme activity, we propose a model for the control of AceCS1 by SIRT1 and of AceCS2 by SIRT3.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
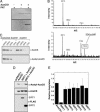
Fig. 1.
AceCS1 is acetylated in vitro and in mammalian cells. (A) AceCS1 is acetylated in vitro by PAT. Recombinant AceCS1 was incubated in the presence or absence of PAT and [1-14C]acetyl-CoA for 1h, resolved by SDS/PAGE and detected by Coomassie (Upper) and autoradiography (Lower). (B) MALDI-TOF confirms Lys-661 is acetylated on AceCS1. (Upper) Tryptic digest of AceCS1. (Lower) tryptic digest of acetylated AceCS1. As expected, the spectra are similar, with Lower showing a new peak corresponding to the acetylated peptide, SGK(ac)IMR. This peptide was further confirmed by MS/MS on the TOF–TOF instrument. (C) Acetylation state of AceCS1 can be detected with an anti-acetyl-AceCS antibody. Recombinant AceCS1 and acetyl-AceCS1 were resolved by SDS/PAGE and detected by Western blotting with anti-AceCS and anti-acetyl-AceCS antibodies, respectively. (D) Acetylation of AceCS1 is decreased upon SIRT1 coexpression. Cos-7 cells cotransfected with a construct expressing AceCS1 and the construct expressing SIRT1 or SIRT2. After 48 h, cell extracts were resolved by SDS/PAGE and detected by Western blot, using anti-acetyl-AceCS, anti-AceCS, and anti-FLAG antibodies. (E) Coexpression of AceCS1 and SIRT1–7 show increased deactylation of AceCS1 by SIRT1. The ratio of acetylated AceCS1 to total AceCS1 from cell lysates was calculated from densitometry of Western blots with anti-AceCS and anti-acetyl-AceCS1 antibodies relative to AceCS1 transfection alone.
Fig. 2.
AceCS1 activity is regulated by sirtuin-catalyzed deacetylation. (A) AceCS1 activity is regulated by reversible acetylation. AceCS1 or acetylated AceCS1 (5 μM) was incubated for 1 h with 2 μM SIRT1 at 37°C. AceCS1 activity was determined by measuring the formation of acetyl-CoA over time from [14C]acetate, and reported as nmol of acetyl-CoA per min per mg of protein. (B) 1,2-[14C]acetate incorporation into lipids through AceCS1 is enhanced upon SIRT1 coexpression. Cos-7 cells were cotransfected with a vector encoding AceCS1 and/or SIRT1, SIRT2, or empty vector where indicated. Lipids were extracted as described in Materials and Methods, and [14C]acetate incorporation was analyzed by scintillation counting and normalized to protein content. ANOVA statistical analysis was performed, indicating significant differences (P < 0.05) between SIRT1 and AceCS1 coexpression and AceCS1 expression alone. (C) SIRT1 and SIRT3 show high catalytic activity against acetylated AceCS1. Catalytic amounts of SIRT1 (filled circles), SIRT2 (open circles), SIRT3 (filled squares), SIRT5 (filled diamonds), and SIRT6 (open squares) (50 nM) were incubated with 1.5 μM acetylated AceCS1 in 1 mM NAD+, 1 mM DTT, and 50 mM Tris (pH 7.5) at 37°C for the indicated time and deacetylation of AceCS1 was measured by AceCS1 activity assays. Data from SIRT1 and SIRT3 were fitted to the integrated Michaelis–Menten equation (45).
Fig. 3.
AceCS2 activity is regulated by SIRT3-catalyzed deacetylation. (A) AceCS2 is acetylated in vitro by PAT. Recombinant AceCS2 was incubated in the presence or absence of PAT and [14C]acetyl-CoA for 1 h, resolved by SDS/PAGE, and detected by Coomassie (Upper) and autoradiography (Lower). (B) AceCS2 activity is regulated by reversible acetylation. AceCS2 or acetylated AceCS2 (2 μM) activity was determined by measuring the formation of acetyl-CoA over time from [14C]acetate, and reported as nmol of acetyl-CoA per min per mg of protein. (C) AceCS2 is preferentially deacetylated by SIRT3. Catalytic amounts of SIRT1, SIRT2, and SIRT3 (50 nM) were incubated with 1.0 μM acetylated AceCS2 in 1 mM NAD+, 1 mM DTT, and 50 mM Tris (pH 7.5) at 37°C with time points taken over 40 min. The ability of sirtuins to deacetylate and activate acetylated AceCS2 was followed by measuring the rate acetyl-CoA formation (nmol of acetyl-CoA per min per mg of protein). (D) Acetylation state of AceCS2 can be detected with an anti-acetyl-AceCS antibody. Recombinant AceCS2 and acetyl-AceCS2 were resolved by SDS/PAGE and detected by Western blotting with anti-AceCS and anti-acetyl-AceCS antibodies, respectively. (E) MALDI-TOF confirms Lys-635 is acetylated on AceCS2. (Upper) Tryptic digest of AceCS2. (Lower) Tryptic digest of acetylated AceCS2. As expected, the spectra are similar, with Lower showing a new peak corresponding to the acetylated peptide, SGK(ac)VMR. This peptide was further confirmed by MS/MS on the TOF-TOF instrument.
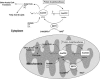
Fig. 4.
Proposed model for the regulation of mammalian AceCSs AceCS1 and AceCS2 by sirtuins SIRT1 and SIRT3. AceCSs are inactivated by protein acetyltransferases (PATs), which acetylate an important catalytic lysine residue. The PAT(s) responsible for acetylating AceCSs remain to be determined. In the cytoplasm, SIRT1 catalyzes the deacetylation of AceCS1 on Lys-661 (murine). In the mitochondria, SIRT3 catalyzes the deacetylation of AceCS2 on Lys-635 (murine). Free acetate, generated from endogenous cellular reactions or absorbed from the gut, is converted to acetyl-CoA that can be used in metabolic pathways, such as fatty acid synthesis (cytoplasm) or the tricarboxylic acid cycle in mitochondria. Other acetyl-CoA requiring enzymes/pathways also may be regulated by sirtuin-controlled AceCS activity.
Similar articles
- Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2.
Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Schwer B, et al. Proc Natl Acad Sci U S A. 2006 Jul 5;103(27):10224-10229. doi: 10.1073/pnas.0603968103. Epub 2006 Jun 20. Proc Natl Acad Sci U S A. 2006. PMID: 16788062 Free PMC article. - SIRT1 and SIRT3 deacetylate homologous substrates: AceCS1,2 and HMGCS1,2.
Hirschey MD, Shimazu T, Capra JA, Pollard KS, Verdin E. Hirschey MD, et al. Aging (Albany NY). 2011 Jun;3(6):635-42. doi: 10.18632/aging.100339. Aging (Albany NY). 2011. PMID: 21701047 Free PMC article. - Acetate metabolism and aging: An emerging connection.
Shimazu T, Hirschey MD, Huang JY, Ho LT, Verdin E. Shimazu T, et al. Mech Ageing Dev. 2010 Jul-Aug;131(7-8):511-6. doi: 10.1016/j.mad.2010.05.001. Epub 2010 May 15. Mech Ageing Dev. 2010. PMID: 20478325 - A link between transcription and intermediary metabolism: a role for Sir2 in the control of acetyl-coenzyme A synthetase.
Starai VJ, Takahashi H, Boeke JD, Escalante-Semerena JC. Starai VJ, et al. Curr Opin Microbiol. 2004 Apr;7(2):115-9. doi: 10.1016/j.mib.2004.02.005. Curr Opin Microbiol. 2004. PMID: 15063846 Review. - Mitochondrial sirtuins.
Huang JY, Hirschey MD, Shimazu T, Ho L, Verdin E. Huang JY, et al. Biochim Biophys Acta. 2010 Aug;1804(8):1645-51. doi: 10.1016/j.bbapap.2009.12.021. Epub 2010 Jan 7. Biochim Biophys Acta. 2010. PMID: 20060508 Review.
Cited by
- Metabolic mechanisms orchestrated by Sirtuin family to modulate inflammatory responses.
Li X, Li Y, Hao Q, Jin J, Wang Y. Li X, et al. Front Immunol. 2024 Sep 20;15:1448535. doi: 10.3389/fimmu.2024.1448535. eCollection 2024. Front Immunol. 2024. PMID: 39372420 Free PMC article. Review. - A Multi-Target Pharmacological Correction of a Lipoyltransferase LIPT1 Gene Mutation in Patient-Derived Cellular Models.
Gómez-Fernández D, Romero-González A, Suárez-Rivero JM, Cilleros-Holgado P, Álvarez-Córdoba M, Piñero-Pérez R, Romero-Domínguez JM, Reche-López D, López-Cabrera A, Ibáñez-Mico S, Castro de Oliveira M, Rodríguez-Sacristán A, González-Granero S, García-Verdugo JM, Sánchez-Alcázar JA. Gómez-Fernández D, et al. Antioxidants (Basel). 2024 Aug 22;13(8):1023. doi: 10.3390/antiox13081023. Antioxidants (Basel). 2024. PMID: 39199267 Free PMC article. - The Role of Sirtuin-1 Isoforms in Regulating Mitochondrial Function.
Patyal P, Ameer FS, Verma A, Zhang X, Azhar G, Shrivastava J, Sharma S, Zhang R, Wei JY. Patyal P, et al. Curr Issues Mol Biol. 2024 Aug 14;46(8):8835-8851. doi: 10.3390/cimb46080522. Curr Issues Mol Biol. 2024. PMID: 39194739 Free PMC article. - The role of SIRT3 in homeostasis and cellular health.
Trinh D, Al Halabi L, Brar H, Kametani M, Nash JE. Trinh D, et al. Front Cell Neurosci. 2024 Aug 2;18:1434459. doi: 10.3389/fncel.2024.1434459. eCollection 2024. Front Cell Neurosci. 2024. PMID: 39157755 Free PMC article. Review. - Acetyl-CoA synthetase activity is enzymatically regulated by lysine acetylation using acetyl-CoA or acetyl-phosphate as donor molecule.
Qin C, Graf LG, Striska K, Janetzky M, Geist N, Specht R, Schulze S, Palm GJ, Girbardt B, Dörre B, Berndt L, Kemnitz S, Doerr M, Bornscheuer UT, Delcea M, Lammers M. Qin C, et al. Nat Commun. 2024 Jul 17;15(1):6002. doi: 10.1038/s41467-024-49952-0. Nat Commun. 2024. PMID: 39019872 Free PMC article.
References
- Rusche L. N., Kirchmaier A. L., Rine J. Annu. Rev. Biochem. 2003;72:481–516. - PubMed
- Gasser S. M., Cockell M. M. Gene. 2001;279:1–16. - PubMed
- Starai V. J., Celic I., Cole R. N., Boeke J. D., Escalante-Semerena J. C. Science. 2002;298:2390–2392. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM065386/GM/NIGMS NIH HHS/United States
- R01 GM065386/GM/NIGMS NIH HHS/United States
- GM059785/GM/NIGMS NIH HHS/United States
- R37 GM059785/GM/NIGMS NIH HHS/United States
- R01 GM059785/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials