Identification of markers for Helicobacter pylori strains isolated from children with peptic ulcer disease by suppressive subtractive hybridization - PubMed (original) (raw)
Identification of markers for Helicobacter pylori strains isolated from children with peptic ulcer disease by suppressive subtractive hybridization
Mónica Oleastro et al. Infect Immun. 2006 Jul.
Abstract
Peptic ulcer disease (PUD) occurs after a long-term Helicobacter pylori infection. However, the disease can develop earlier, and rare cases have been observed in children, suggesting that these H. pylori strains may be more virulent. We used suppressive subtractive hybridization for comparative genomics between H. pylori strains isolated from a 5-year-old child with duodenal ulcer and from a sex- and age-matched child with gastritis only. The prevalence of the 30 tester-specific subtracted sequences was determined on a collection of H. pylori strains from children (15 ulcers and 30 gastritis) and from adults (46 ulcers and 44 gastritis). Two of these sequences, jhp0562 (80.0% versus 33.3%, P = 0.008) and jhp0870 (80.0% versus 36.7%, P = 0.015), were highly associated with PUD in children and a third sequence, jhp0828, was less associated (40.0% versus 10.0%, P = 0.048). Among adult strains, none of the 30 sequences was associated with PUD. However, both jhp0562 and jhp0870 were less prevalent in adenocarcinoma strains than in PUD strains from children and adults, the difference being statistically significant for jhp0870. In conclusion, two H. pylori genes were identified as being strongly associated with PUD in children, and their putative roles as an outer membrane protein for jhp0870 and in lipopolysaccharide biosynthesis for jhp0562, suggest that they may be novel virulence factors of H. pylori.
Figures
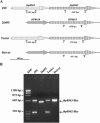
FIG. 1.
Schematic representation of the PCR assay for the detection of the jhp0562, jhp0563(HP0619)-like genes in H. pylori strains (A) and analysis of their detection by PCR in an ethidium bromide-stained 1.5% agarose gel (B). (A) The black arrows represent the forward F1-jhp0562/jhp0563, labeled F, and the reverse R1-jhp0562/jhp0563, labeled R, conserved primers between H. pylori strains, respectively. These F and R primers hybridize both on jhp0562 and on jhp0563-like genes. Genes that belong to the same family are represented in an identical way. (B) MW, molecular weight marker ΦX174 RF DNA/HaeIII fragments (72-1353 pb; Invitrogen).
FIG. 2.
Genomic organization of the jhp0870 (A) and jhp0649/HP0710 (B) loci in J99 and 26695 H. pylori strains. The jhp0870 gene is specific for the J99 strain. No homologue of this gene exists in the 26695 strain (5). Instead, an intergenic space is observed, flanked by genes homologous to those flanking jhp0870 in the J99 strain (52). Genes which belong to the same family are represented in an identical way. The boxes represent the 27-, 2-, and 4-bp additional sequences present in jhp0870 but absent in jhp0649/HP0710.
FIG. 3.
Schematic representation of the PCR assay for the detection of the jhp0870 and jhp0649-like genes in H. pylori strains (A) and analysis of their detection by PCR in ethidium bromide-stained 2,5% agarose gel (B). (A) The black arrows represent the forward F1-jhp0870/jhp0649, labeled F, and reverse F1-jhp0870/jhp0649, labeled R, conserved primers between the jhp0870 and jhp0649-like genes in H. pylori strains, respectively. These F and R primers hybridize both on jhp0649 and jhp0870-like genes. The three internal boxes in the jhp0870 scheme represent the 27-bp, 2-bp and 4-bp additional sequences present in jhp0870 but absent in jhp0649. (B) MW, molecular weight marker ΦX174 RF DNA/HaeIII fragments (72-1353 pb; Invitrogen).
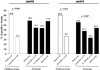
FIG. 4.
Distribution of jhp0562 and jhp0870 genes among H. pylori strains isolated from subjects with PUD, gastritis, gastric adenocarcinoma, and gastric MALT lymphoma, including both children only and children and adults together (all strains). P values, determined by the Fisher exact test, are shown for the significant associations only.
Similar articles
- Prevalence of Helicobacter pylori cagA, vacA, iceA, babA2 genotypes in Polish children and adolescents with gastroduodenal disease.
Biernat MM, Gościniak G, Iwańczak B. Biernat MM, et al. Postepy Hig Med Dosw (Online). 2014 Aug 22;68:1015-21. doi: 10.5604/17322693.1118211. Postepy Hig Med Dosw (Online). 2014. PMID: 25228509 - Equally high prevalences of infection with cagA-positive Helicobacter pylori in Chinese patients with peptic ulcer disease and those with chronic gastritis-associated dyspepsia.
Pan ZJ, van der Hulst RW, Feller M, Xiao SD, Tytgat GN, Dankert J, van der Ende A. Pan ZJ, et al. J Clin Microbiol. 1997 Jun;35(6):1344-7. doi: 10.1128/jcm.35.6.1344-1347.1997. J Clin Microbiol. 1997. PMID: 9163441 Free PMC article. - Helicobacter pylori cagA status and peptic ulcer disease in Iran.
Salehi Z, Jelodar MH, Rassa M, Ahaki M, Mollasalehi H, Mashayekhi F. Salehi Z, et al. Dig Dis Sci. 2009 Mar;54(3):608-13. doi: 10.1007/s10620-008-0378-8. Epub 2008 Jul 10. Dig Dis Sci. 2009. PMID: 18612816 - Peptic ulcer and Helicobacter pylori.
Cohen H. Cohen H. Gastroenterol Clin North Am. 2000 Dec;29(4):775-89. doi: 10.1016/s0889-8553(05)70146-1. Gastroenterol Clin North Am. 2000. PMID: 11190063 Review. - [Peptic ulcer and Helicobacter pyrlori. Results and consequences of its eradication].
Ballesteros-Amozurrutia MA. Ballesteros-Amozurrutia MA. Rev Gastroenterol Mex. 2000 Oct-Dec;65(4 Suppl 2):41-9. Rev Gastroenterol Mex. 2000. PMID: 11464621 Review. Spanish.
Cited by
- Ulcerogenic Helicobacter pylori strains isolated from children: a contribution to get insight into the virulence of the bacteria.
Vitoriano I, Saraiva-Pava KD, Rocha-Gonçalves A, Santos A, Lopes AI, Oleastro M, Roxo-Rosa M. Vitoriano I, et al. PLoS One. 2011;6(10):e26265. doi: 10.1371/journal.pone.0026265. Epub 2011 Oct 19. PLoS One. 2011. PMID: 22039453 Free PMC article. - The Role of Helicobacter pylori Outer Membrane Proteins in Adherence and Pathogenesis.
Oleastro M, Ménard A. Oleastro M, et al. Biology (Basel). 2013 Aug 27;2(3):1110-34. doi: 10.3390/biology2031110. Biology (Basel). 2013. PMID: 24833057 Free PMC article. - Surveillance of Helicobacter pylori Antibiotic Susceptibility in Indonesia: Different Resistance Types among Regions and with Novel Genetic Mutations.
Miftahussurur M, Syam AF, Nusi IA, Makmun D, Waskito LA, Zein LH, Akil F, Uwan WB, Simanjuntak D, Wibawa ID, Waleleng JB, Saudale AM, Yusuf F, Mustika S, Adi P, Maimunah U, Maulahela H, Rezkitha YA, Subsomwong P, Nasronudin, Rahardjo D, Suzuki R, Akada J, Yamaoka Y. Miftahussurur M, et al. PLoS One. 2016 Dec 1;11(12):e0166199. doi: 10.1371/journal.pone.0166199. eCollection 2016. PLoS One. 2016. PMID: 27906990 Free PMC article. - Novel functions for glycosyltransferases Jhp0562 and GalT in Lewis antigen synthesis and variation in Helicobacter pylori.
Pohl MA, Kienesberger S, Blaser MJ. Pohl MA, et al. Infect Immun. 2012 Apr;80(4):1593-605. doi: 10.1128/IAI.00032-12. Epub 2012 Jan 30. Infect Immun. 2012. PMID: 22290141 Free PMC article. - Biomarker Characterization and Prediction of Virulence and Antibiotic Resistance from Helicobacter pylori Next Generation Sequencing Data.
Vital JS, Tanoeiro L, Lopes-Oliveira R, Vale FF. Vital JS, et al. Biomolecules. 2022 May 11;12(5):691. doi: 10.3390/biom12050691. Biomolecules. 2022. PMID: 35625618 Free PMC article. Review.
References
- Agron, P. G., M. Macht, L. Radnedge, E. W. Skowronski, W. Miller, and G. L. Andersen. 2002. Use of subtractive hybridization for comprehensive surveys of prokaryotic genome differences. FEMS Microbiol. Lett. 211:175-182. - PubMed
- Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-53. - PubMed
- Alarcon, T., D. Domingo, M. J. Martinez, and M. LopezBrea. 1999. cagA gene and vacA alleles in Spanish Helicobacter pylori clinical isolates from patients of different ages. FEMS Immunol. Med. Microbiol. 24:215-219. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases