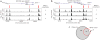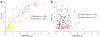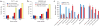DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays - PubMed (original) (raw)
DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays
Gregory E Crawford et al. Nat Methods. 2006 Jul.
Abstract
Mapping DNase I hypersensitive sites is an accurate method of identifying the location of gene regulatory elements, including promoters, enhancers, silencers and locus control regions. Although Southern blots are the traditional method of identifying DNase I hypersensitive sites, the conventional manual method is not readily scalable to studying large chromosomal regions, much less the entire genome. Here we describe DNase-chip, an approach that can rapidly identify DNase I hypersensitive sites for any region of interest, or potentially for the entire genome, by using tiled microarrays. We used DNase-chip to identify DNase I hypersensitive sites accurately from a representative 1% of the human genome in both primary and immortalized cell types. We found that although most DNase I hypersensitive sites were present in both cell types studied, some of them were cell-type specific. This method can be applied globally or in a targeted fashion to any tissue from any species with a sequenced genome.
Figures
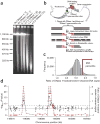
Figure 1. DNase-chip protocol
(a) Pulsed field gel electrophoresis of DNase I–digested nuclear DNA. The concentrations of DNase I used for DNase-chip are labeled as A, B and C. (b) Outline of DNase-chip protocol. (c) Histogram of signal ratios of DNase I–treated versus random-sheared DNA. Tiled oligos that displayed the top 5% ratios are located to the right of the red bar. (d) Identification of regions with significant P values. The raw ratio data are plotted in gray, with the y-axis label on the right; the top 5% cutoff is displayed as a dotted horizontal gray line. The P value data for sliding 500-bp windows are plotted in red, with the _y_-axis label on the left.
Figure 2. Confirmatory real-time PCR analysis
(a–f) DNase-chip–identified regions were confirmed by real-time PCR for CD4+ T cells (a,c,e) and GM06990 cells (b,d,f). ΔCt values represent the number of additional cycles to achieve threshold levels of amplification between DNase I–treated and non-digested nuclear DNA. Higher ΔCt values represent elevated levels of DNase I–digestion. Control primer sets were designed around random regions of the genome, as well as known DNase I hypersensitive sites generated from a separate study (MPSS cluster). Real-time PCR using primer sets flanking DNase-chip peaks that are present with all three DNase I concentrations (a,b). Real-time PCR using primers sets flanking DNase-chip peaks that are present with two out of three DNase I concentrations (c,d). Real-time PCR using primer sets flanking DNase-chip peaks that are present with only a single DNase I concentration (e,f).
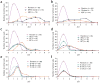
Figure 3. Comparison of DNase-chip and MPSS data for CD4+ T cells
(a,b) DNase-chip P value data from two different ENCODE regions shows peaks that are detected using multiple concentrations of DNase I. MPSS clusters (in red) show DNase I hypersensitive sites that were previously identified using a sequence-based approach and clustering of tags. Note that there are regions detected by DNase-chip that have not been detected by MPSS, as expected because of limitations in the number of sequence tags. Many DNase-chip peaks that are only detected with a single DNase I concentration appear to be false positives. (c) Venn diagram shows the number of DNase I hypersensitive sites identified by DNase-chip (black; the number of DNase-chip regions are normalized using positive predictive values) and validated MPSS clusters (red).
Figure 4. Identification of cell type–specific DNase I hypersensitive sites
(a,b) Real-time PCR was performed on both CD4+ T cells and GM06990 cells. Real-time PCR using primer sets that flank random regions of the genome or DNase-chip peaks that are present for both cell types (a). Real-time PCR using primer sets that flank DNase-chip peaks that are present in only CD4+ T cells (CD4) or GM06990 cells (GM; b).
Figure 5. Location of DNase I hypersensitive sites relative to the annotated genome
(a,b) DNase-chip peaks were mapped to ENCODE regions stratified by gene density and human-mouse sequence conservation for both CD4+ T cells (a) and the GM06990 lymphoblastoid cell line (b). (c) The genomic locations of DNase I hypersensitive sites (detected with at least two concentrations of DNase I) and computationally generated random controls (n = 1,000) were compared to Gencode transcription start and end sites (within a 2-kb window), CpG islands, first introns, non-first introns, first exons, non-first exons, conserved sequences (MCS), conserved sequences minus coding exons (MCS-no-CDS). The number of DNase I hypersensitive sites at different distances (0 kb, 2 kb, 10 kb, and 25 kb) from Gencode genes was also determined. Error bars represent the entire range of values seen randomly generated mock datasets (n = 1,000). Compared to the random controls, the locations of the DNase-chip peaks are significantly (Monte Carlo P < 0.001) over- or under-represented at all positions except transcription end and > 0 kb from gene.
Figure 6. Expression of genes relative to proximity to DNase I hypersensitive sites
(a,b) The distance of each transcription start site (blue dots) to the nearest DNase I hypersensitive site was compared to the gene expression values of each transcript for both CD4+ T cells (a) and the GM06990 lymphoblastoid cell line (b). Horizontal red lines mark the expression level that separates most genes that have a DNase I hypersensitive site nearby (<1,000 bp) versus those that do not (>1,000 bp). (c) Average expression values of genes that have a DNase I hypersensitive site within 1 kb versus those that do not were determined for both cell types. Average expression values of genes that have a DNase I hypersensitive site are significantly different from genes that do not (P < 1 × 10−38). CD4+ T cells, CD4; or GM06990 cells, GM.
Comment in
- How to find an opening (or lots of them).
Giresi PG, Lieb JD. Giresi PG, et al. Nat Methods. 2006 Jul;3(7):501-2. doi: 10.1038/nmeth0706-501. Nat Methods. 2006. PMID: 16791206 No abstract available. - Hacking the genome.
Evanko D. Evanko D. Nat Methods. 2006 Jul;3(7):495. doi: 10.1038/nmeth0706-495. Nat Methods. 2006. PMID: 16832861 No abstract available.
Similar articles
- Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays.
Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, Weaver M, Shafer A, Lee K, Neri F, Humbert R, Singer MA, Richmond TA, Dorschner MO, McArthur M, Hawrylycz M, Green RD, Navas PA, Noble WS, Stamatoyannopoulos JA. Sabo PJ, et al. Nat Methods. 2006 Jul;3(7):511-8. doi: 10.1038/nmeth890. Nat Methods. 2006. PMID: 16791208 - Mapping regulatory elements by DNaseI hypersensitivity chip (DNase-Chip).
Shibata Y, Crawford GE. Shibata Y, et al. Methods Mol Biol. 2009;556:177-90. doi: 10.1007/978-1-60327-192-9_13. Methods Mol Biol. 2009. PMID: 19488879 - High-resolution mapping and characterization of open chromatin across the genome.
Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. Boyle AP, et al. Cell. 2008 Jan 25;132(2):311-22. doi: 10.1016/j.cell.2007.12.014. Cell. 2008. PMID: 18243105 Free PMC article. - Advances of DNase-seq for mapping active gene regulatory elements across the genome in animals.
Chen A, Chen D, Chen Y. Chen A, et al. Gene. 2018 Aug 15;667:83-94. doi: 10.1016/j.gene.2018.05.033. Epub 2018 May 14. Gene. 2018. PMID: 29772251 Review. - Open chromatin in plant genomes.
Zhang W, Zhang T, Wu Y, Jiang J. Zhang W, et al. Cytogenet Genome Res. 2014;143(1-3):18-27. doi: 10.1159/000362827. Epub 2014 Jun 6. Cytogenet Genome Res. 2014. PMID: 24923879 Review.
Cited by
- DNase I digestion of isolated nulcei for genome-wide mapping of DNase hypersensitivity sites in chromatin.
Ling G, Waxman DJ. Ling G, et al. Methods Mol Biol. 2013;977:21-33. doi: 10.1007/978-1-62703-284-1_3. Methods Mol Biol. 2013. PMID: 23436351 Free PMC article. - Genome-Wide Identification of Regulatory Sequences Undergoing Accelerated Evolution in the Human Genome.
Dong X, Wang X, Zhang F, Tian W. Dong X, et al. Mol Biol Evol. 2016 Oct;33(10):2565-75. doi: 10.1093/molbev/msw128. Epub 2016 Jul 8. Mol Biol Evol. 2016. PMID: 27401230 Free PMC article. - Characterization of enhancer activity in early human neurodevelopment using Massively Parallel Reporter Assay (MPRA) and forebrain organoids.
Capauto D, Wang Y, Wu F, Norton S, Mariani J, Inoue F, Crawford GE, Ahituv N, Abyzov A, Vaccarino FM. Capauto D, et al. Sci Rep. 2024 Feb 16;14(1):3936. doi: 10.1038/s41598-024-54302-7. Sci Rep. 2024. PMID: 38365907 Free PMC article. - Mapping accessible chromatin regions using Sono-Seq.
Auerbach RK, Euskirchen G, Rozowsky J, Lamarre-Vincent N, Moqtaderi Z, Lefrançois P, Struhl K, Gerstein M, Snyder M. Auerbach RK, et al. Proc Natl Acad Sci U S A. 2009 Sep 1;106(35):14926-31. doi: 10.1073/pnas.0905443106. Epub 2009 Aug 18. Proc Natl Acad Sci U S A. 2009. PMID: 19706456 Free PMC article. - Current concepts of follicle-stimulating hormone receptor gene regulation.
George JW, Dille EA, Heckert LL. George JW, et al. Biol Reprod. 2011 Jan;84(1):7-17. doi: 10.1095/biolreprod.110.085043. Epub 2010 Aug 25. Biol Reprod. 2011. PMID: 20739665 Free PMC article. Review.
References
- The Encode Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. - PubMed
- Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. - PubMed
- Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. - PubMed
- Wu C, Wong YC, Elgin SC. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979;16:807–814. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases