Broadening of DNA replication origin usage during metazoan cell differentiation - PubMed (original) (raw)
Broadening of DNA replication origin usage during metazoan cell differentiation
Sébastien Dazy et al. EMBO Rep. 2006 Aug.
Abstract
We have examined whether replication of the chicken beta-globin locus changes during differentiation of primary erythroid progenitors into erythrocytes. In undifferentiated progenitors, four principal initiation sites and a replication fork pausing region (RFP) were observed. Forty-eight hours after induction of differentiation, the principal sites were maintained, even in the activated beta(A)-globin gene, some minor sites were enhanced, three new sites appeared and the RFP disappeared. One of the activated origins showed increased histone H3 K9K14 diacetylation, but the others did not. These results demonstrate a broadening of DNA replication origin usage during differentiation of untransformed metazoan cells and indicate that histone H3 diacetylation, other histone modifications so far reported and transcription are not crucial determinants of origin selection in this system.
Figures
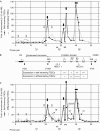
Figure 1
Activation of tissue-specific origins during erythroid differentiation. (A, C) Fold of enrichments of 2.0–3.5 kb (A) and 2.0–3.0 kb (C) nascent strands (NS) extracted from two independently prepared batches of either self-renewing T2ECs (black circles) or 48 h differentiating T2ECs (grey triangles) and quantified by real-time PCR. Each quantification was repeated at least twice. Error bars indicate standard deviation for samples quantified four or six times. The scale corresponds to fold of enrichments over the background. The abscissa scale is map position (nucleotide number). Black and grey arrows in the graph point towards constitutive and differentiation-activated origins, respectively. The horizontal double arrow over the βA-globin gene indicates a 1.5 kb zone of initiation. The position of a bidirectional replication fork pausing site inside the ρ-globin gene in self-renewing T2ECs is shown below each graph (double black arrowheads). The positions of primer pairs 10, 20 and 30 are shown further below. Additional primer pairs (18′, 23′, 36, 37, 38 and 39) were used for the differentiation shown in (C). The position of primer pairs 18′, 23′, 27 and 31 is shown inside the graph. (B) Map of the folate receptor gene, condensed chromatin region, β-globin domain and olfactory receptor gene (COR3′). DNase I hypersensitive sites (HSs; HSA, HS1–3, HS βA/ɛ, 5′HS4 and 3′HS) are indicated. Expression of globin genes in self-renewing and differentiating T2ECs is indicated below the map. +/− and ++++ indicate weak and very strong expression, respectively.
Figure 2
Nascent strand maturation analysis around the ρ- and βA-globin genes. Fold of enrichments of 0.8–1.2 kb (black triangles) and 2.0–3.0 kb (grey triangles) nascent strands (NS) extracted from the same batch of differentiating T2ECs as in Fig 1B and quantified with primer pairs inside and around the βA-gene (A) or the ρ-gene (B). Each quantification was performed twice. Below the graph, the coding regions of ρ- and βA-globin genes are shown. Grey rectangles, exons 1–3; black rectangle, βA/ɛ enhancer; black bars, amplicons used for real-time PCR quantifications. Grey bars inside graphs delineate 1 kb regions devoid of origin activity because of being centred on a primer pair that gives enrichment close to background level. Vertical and horizontal black arrows indicate origins and initiation zones, respectively. Parts (A, B) demonstrate continued initiation in the βA-gene and downregulation of the ρ-gene replication fork barrier, respectively, in differentiating cells.
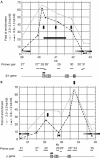
Figure 3
Changes in histone H3 diacetylation during differentiation and comparison with replication changes. Relative recovery of crosslinked protein–DNA complexes recovered after immunoprecipitation with anti-K9K14 diacetylated histone H3 antibody from self-renewing T2ECs (black circles) or 48 h differentiating T2ECs (grey triangles) as determined by real-time PCR with 27 primer pairs (1–7 and 13–33). Results with two independent batches of cells are shown (A, B). Each data point is the mean of two different immunoprecipitations from the same sample of crosslinked chromatin. The ordinate is a relative recovery (% Ac/K9 and K14 H3) normalized with respect to primer pair 5 (arbitrarily set as 100%). The abscissa is map position (nucleotide number). Black and grey arrows in the graph point towards constitutive- and differentiation-activated origins of replication, respectively. The star above one grey arrow indicates a differentiation-activated origin localized in a region where acetylation increases during differentiation. The position of the bidirectional replication fork pausing site inside the ρ-globin gene of undifferentiated cells is indicated (double black arrowheads). FR, folate receptor; HS, hypersensitive site.
Comment in
- DNA replication: the unbearable lightness of origins.
Norio P. Norio P. EMBO Rep. 2006 Aug;7(8):779-81. doi: 10.1038/sj.embor.7400766. EMBO Rep. 2006. PMID: 16880822 Free PMC article. Review. No abstract available.
Similar articles
- Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus.
Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Litt MD, et al. Science. 2001 Sep 28;293(5539):2453-5. doi: 10.1126/science.1064413. Epub 2001 Aug 9. Science. 2001. PMID: 11498546 - HoxB domain induction silences DNA replication origins in the locus and specifies a single origin at its boundary.
Grégoire D, Brodolin K, Méchali M. Grégoire D, et al. EMBO Rep. 2006 Aug;7(8):812-6. doi: 10.1038/sj.embor.7400758. Epub 2006 Jul 14. EMBO Rep. 2006. PMID: 16845368 Free PMC article. - DNA replication: the unbearable lightness of origins.
Norio P. Norio P. EMBO Rep. 2006 Aug;7(8):779-81. doi: 10.1038/sj.embor.7400766. EMBO Rep. 2006. PMID: 16880822 Free PMC article. Review. No abstract available. - The mammalian beta globin origin of DNA replication.
Aladjem MI. Aladjem MI. Front Biosci. 2004 Sep 1;9:2540-7. doi: 10.2741/1415. Front Biosci. 2004. PMID: 15358579 Review.
Cited by
- Replication stress caused by low MCM expression limits fetal erythropoiesis and hematopoietic stem cell functionality.
Alvarez S, Díaz M, Flach J, Rodriguez-Acebes S, López-Contreras AJ, Martínez D, Cañamero M, Fernández-Capetillo O, Isern J, Passegué E, Méndez J. Alvarez S, et al. Nat Commun. 2015 Oct 12;6:8548. doi: 10.1038/ncomms9548. Nat Commun. 2015. PMID: 26456157 Free PMC article. - Mapping of Replication Origins in the X Inactivation Center of Vole Microtus levis Reveals Extended Replication Initiation Zone.
Sherstyuk VV, Shevchenko AI, Zakian SM. Sherstyuk VV, et al. PLoS One. 2015 Jun 3;10(6):e0128497. doi: 10.1371/journal.pone.0128497. eCollection 2015. PLoS One. 2015. PMID: 26038842 Free PMC article. - Replication stress in Mammalian cells and its consequences for mitosis.
Gelot C, Magdalou I, Lopez BS. Gelot C, et al. Genes (Basel). 2015 May 22;6(2):267-98. doi: 10.3390/genes6020267. Genes (Basel). 2015. PMID: 26010955 Free PMC article. Review. - Peaks cloaked in the mist: the landscape of mammalian replication origins.
Hyrien O. Hyrien O. J Cell Biol. 2015 Jan 19;208(2):147-60. doi: 10.1083/jcb.201407004. J Cell Biol. 2015. PMID: 25601401 Free PMC article. Review. - A new light on DNA replication from the inactive X chromosome.
Aladjem MI, Fu H. Aladjem MI, et al. Bioessays. 2014 Jun;36(6):591-7. doi: 10.1002/bies.201400021. Epub 2014 Apr 6. Bioessays. 2014. PMID: 24706495 Free PMC article. Review.
References
- Danis E, Brodolin K, Menut S, Maiorano D, Girard-Reydet C, Mechali M (2004) Specification of a DNA replication origin by a transcription complex. Nat Cell Biol 6: 721–730 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources